Join GitHub today
GitHub is home to over 28 million developers working together to host and review code, manage projects, and build software together.
Sign upInChI generation error #20
Comments
This comment has been minimized.
This comment has been minimized.
schymane
commented
Jan 7, 2019
|
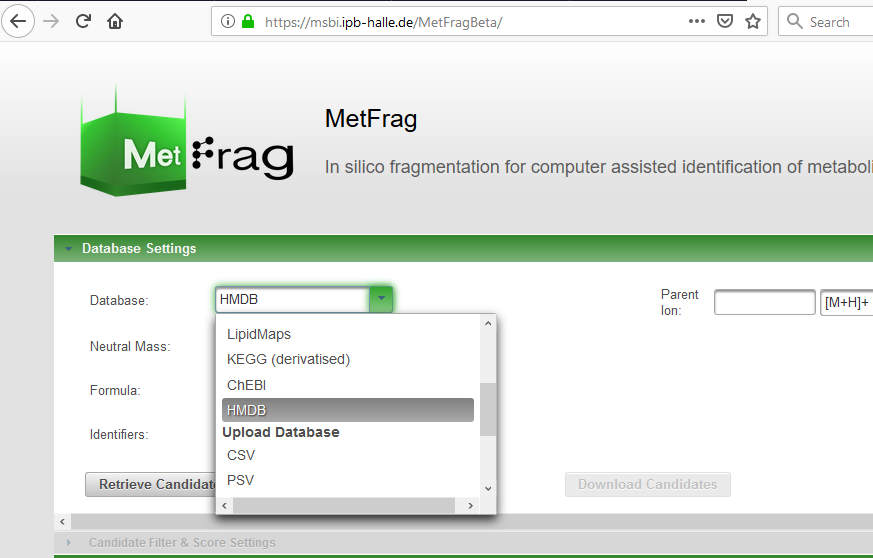
Thanks for the report - are you able to provide the parameters you are using (parameter.txt)? Note also that HMDB is already integrated as a database (second screenshot), so you should be able to access this already using different settings (please try the web interface and download the parameter files to find the correct settings). |
AimeeD90 commentedJan 2, 2019
Hello,
I have tried to use a LocalSDF database of the latest version of HMDB, but an InChI generation error occurred. The error logs are pasted as follows. Is there any solution for this problem?
Besides, at the end of the letter, I provided a specific example that causing this error. Hope that could help test.
ERROR LOGS:
$ java -jar MetFrag2.4.3-CL.jar parameter.txt
org.openscience.cdk.exception.CDKException: Failed to generate InChI: Unsupported bond type
at org.openscience.cdk.inchi.InChIGenerator.generateInchiFromCDKAtomContainer(InChIGenerator.java:307)
at org.openscience.cdk.inchi.InChIGenerator.(InChIGenerator.java:172)
at org.openscience.cdk.inchi.InChIGenerator.(InChIGenerator.java:130)
at org.openscience.cdk.inchi.InChIGeneratorFactory.getInChIGenerator(InChIGeneratorFactory.java:147)
at de.ipbhalle.metfraglib.additionals.MoleculeFunctions.getInChIInfoFromAtomContainer(MoleculeFunctions.java:235)
at de.ipbhalle.metfraglib.database.LocalSDFDatabase.readCandidatesFromFile(LocalSDFDatabase.java:148)
at de.ipbhalle.metfraglib.database.LocalSDFDatabase.getCandidateIdentifiers(LocalSDFDatabase.java:31)
at de.ipbhalle.metfraglib.process.CombinedMetFragProcess.retrieveCompounds(CombinedMetFragProcess.java:77)
at de.ipbhalle.metfrag.commandline.CommandLineTool.main(CommandLineTool.java:104)
java.lang.NullPointerException
at de.ipbhalle.metfraglib.additionals.MoleculeFunctions.getInChIInfoFromAtomContainer(MoleculeFunctions.java:239)
at de.ipbhalle.metfraglib.database.LocalSDFDatabase.readCandidatesFromFile(LocalSDFDatabase.java:148)
at de.ipbhalle.metfraglib.database.LocalSDFDatabase.getCandidateIdentifiers(LocalSDFDatabase.java:31)
at de.ipbhalle.metfraglib.process.CombinedMetFragProcess.retrieveCompounds(CombinedMetFragProcess.java:77)
at de.ipbhalle.metfrag.commandline.CommandLineTool.main(CommandLineTool.java:104)
ERROR de.ipbhalle.metfrag.commandline.CommandLineTool - Error when retrieving compounds.
EXAMPLE:
Mrv0541 04191211592D
95106 0 0 1 0 999 V2000
-0.6472 -1.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7591 -0.7620 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.9518 -1.3667 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6231 -1.8463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3741 -1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4538 -0.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7591 0.9361 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.6472 1.7396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3596 2.1164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4780 2.9329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8302 3.4437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9487 4.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7149 4.5657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9518 1.5366 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.5935 2.0551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7217 1.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3634 1.7586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1333 1.4621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7940 1.7396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9183 0.9361 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1.7425 0.7870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1566 0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7425 -0.6171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9183 -0.7620 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
0.7940 -1.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5313 -1.9465 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6536 -2.7624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4214 -3.0643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5437 -3.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3114 -4.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1235 -1.3667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7616 -1.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8954 -1.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1235 1.5366 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7961 2.0143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5462 1.6707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2188 2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1413 2.9697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5313 2.1164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2821 2.4583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6106 2.9376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3614 3.2795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4408 4.1007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0900 2.1495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0900 2.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0900 -1.9797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0900 -2.8047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5708 -0.6171 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.3677 -0.4036 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-1.5708 0.8118 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.3677 0.5983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0741 -3.1840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7885 -3.5965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5030 -3.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7885 -4.4215 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-3.5030 -4.8340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5030 -5.6590 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-4.3280 -5.6590 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-2.0741 -2.3590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3596 -1.9465 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3596 -2.7715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3008 4.7709 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.0329 2.8002 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
4.9688 1.8048 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1.8983 -4.3941 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-3.2353 2.5736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-4.0453 -1.9845 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.5001 -5.6865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6751 -7.1154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6751 -5.6865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3694 -5.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2626 -6.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1231 -5.0734 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
0.4556 -6.2294 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.9126 -6.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7376 -6.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5001 -7.1154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9126 -7.8299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1575 -6.7815 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0712 -7.6020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8249 -7.9375 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.9964 -8.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7811 -8.9994 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9645 -6.6099 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.3000 -5.8563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3770 -7.3244 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1974 -7.4107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8457 -7.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6780 -5.6590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5030 -6.4840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1984 -6.5819 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
-1.4419 -6.6140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0601 0.1366 0.0000 Co 0 0 0 0 0 0 0 0 0 0 0 0
-0.4324 2.9513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0593 3.8036 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
46 1 2 0 0 0 0
1 2 1 0 0 0 0
60 1 1 0 0 0 0
48 2 1 0 0 0 0
2 93 1 0 0 0 0
48 3 1 0 0 0 0
48 50 1 0 0 0 0
3 4 1 1 0 0 0
3 60 1 0 0 0 0
4 5 1 0 0 0 0
5 67 1 0 0 0 0
5 6 2 0 0 0 0
50 7 1 0 0 0 0
50 14 1 0 0 0 0
7 8 2 0 0 0 0
9 8 1 0 0 0 0
8 44 1 0 0 0 0
9 10 1 6 0 0 0
14 9 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 62 1 0 0 0 0
12 13 2 0 0 0 0
14 15 1 6 0 0 0
14 16 1 0 0 0 0
16 17 1 0 0 0 0
17 66 1 0 0 0 0
17 18 2 0 0 0 0
44 19 2 0 0 0 0
19 20 1 0 0 0 0
19 39 1 0 0 0 0
20 21 2 0 0 0 0
21 22 1 0 0 0 0
21 34 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
23 31 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 1 6 0 0 0
26 31 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 65 1 0 0 0 0
29 30 2 0 0 0 0
31 32 1 0 0 0 0
31 33 1 0 0 0 0
34 35 1 6 0 0 0
34 39 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 64 1 0 0 0 0
37 38 2 0 0 0 0
39 40 1 6 0 0 0
39 41 1 1 0 0 0
41 42 1 0 0 0 0
42 63 1 0 0 0 0
42 43 2 0 0 0 0
20 93 8 0 0 0 0
7 93 8 0 0 0 0
24 93 8 0 0 0 0
44 45 1 0 0 0 0
46 47 1 0 0 0 0
48 49 1 1 0 0 0
50 51 1 6 0 0 0
59 52 1 0 0 0 0
52 53 1 0 0 0 0
53 55 1 0 0 0 0
53 54 2 0 0 0 0
56 55 1 0 0 0 0
57 56 1 0 0 0 0
57 89 1 1 0 0 0
57 58 1 1 0 0 0
57 90 1 0 0 0 0
60 59 1 6 0 0 0
60 61 1 1 0 0 0
46 25 1 0 0 0 0
68 70 2 0 0 0 0
68 75 1 0 0 0 0
72 69 2 0 0 0 0
69 77 1 0 0 0 0
70 73 1 0 0 0 0
70 72 1 0 0 0 0
71 74 1 0 0 0 0
71 73 2 0 0 0 0
74 72 1 0 0 0 0
79 74 1 1 0 0 0
77 75 2 0 0 0 0
75 76 1 0 0 0 0
77 78 1 0 0 0 0
79 84 1 0 0 0 0
79 80 1 0 0 0 0
81 80 1 0 0 0 0
86 81 1 0 0 0 0
81 82 1 6 0 0 0
82 83 1 0 0 0 0
84 86 1 0 0 0 0
84 85 1 1 0 0 0
86 87 1 1 0 0 0
91 87 1 0 0 0 0
91 88 1 0 0 0 0
91 89 1 0 0 0 0
91 92 2 0 0 0 0
93 94 1 0 0 0 0
94 95 3 0 0 0 0
73 93 8 0 0 0 0
M STY 4 1 DAT 2 DAT 3 DAT 4 DAT
M SAL 1 2 20 93
M SDT 1 MRV_COORDINATE_BOND_TYPE
M SDD 1 0.0000 0.0000 DR ALL 0 0
M SED 1 59
M SAL 2 2 7 93
M SDT 2 MRV_COORDINATE_BOND_TYPE
M SDD 2 0.0000 0.0000 DR ALL 0 0
M SED 2 60
M SAL 3 2 24 93
M SDT 3 MRV_COORDINATE_BOND_TYPE
M SDD 3 0.0000 0.0000 DR ALL 0 0
M SED 3 61
M SAL 4 2 73 93
M SDT 4 MRV_COORDINATE_BOND_TYPE
M SDD 4 0.0000 0.0000 DR ALL 0 0
M SED 4 106
M END
OC[C@H]1OC@@HN1C=NC2=CC(C)=C(C)C=C12
C63H89CoN14O14P
$$$$