Join GitHub today
GitHub is home to over 36 million developers working together to host and review code, manage projects, and build software together.
Sign up[REVIEW]: NiBetaSeries: task related correlations in fMRI #1295
Comments
 whedon
assigned
arokem
whedon
assigned
arokem
Mar 4, 2019
 whedon
added
the
review
label
whedon
added
the
review
label
Mar 4, 2019
 whedon
referenced this issue
whedon
referenced this issue
Mar 4, 2019
Closed
[PRE REVIEW]: NiBetaSeries: task related correlations in fMRI #1267
This comment has been minimized.
This comment has been minimized.
|
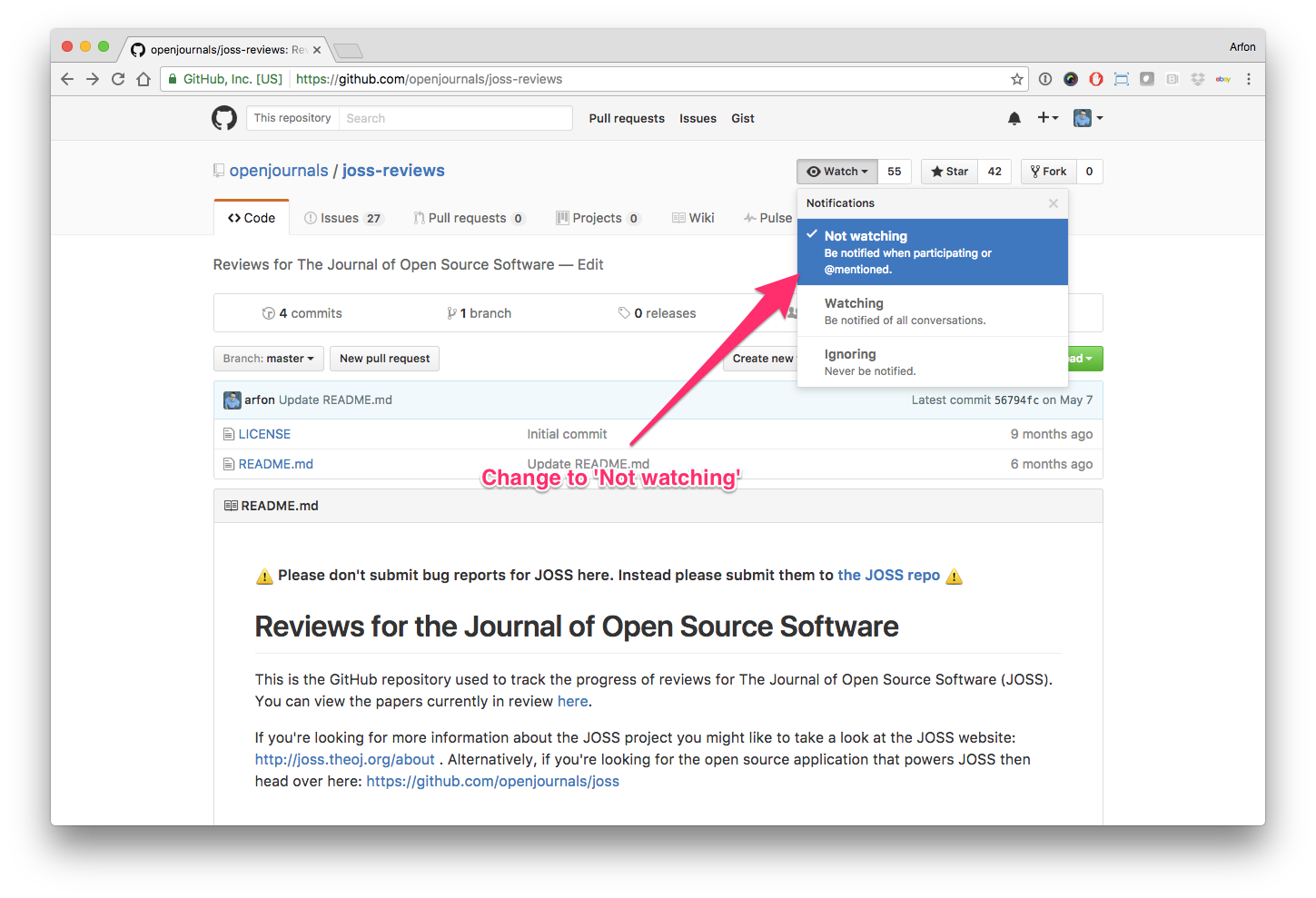
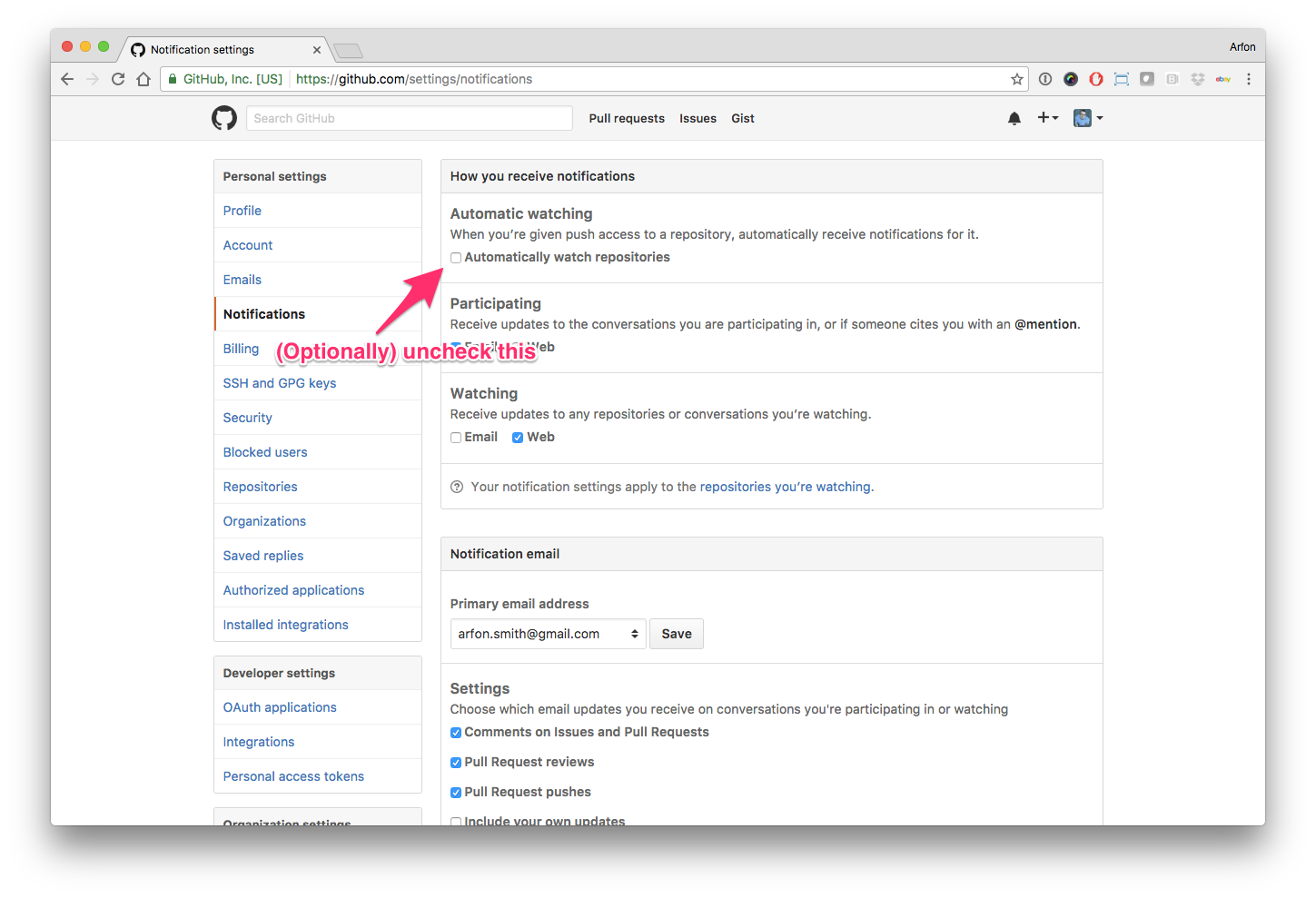
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @snastase it looks like you're currently assigned as the reviewer for this paper If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews To fix this do the following two things:
For a list of things I can do to help you, just type: |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
arokem
commented
Mar 25, 2019
|
@snastase : have you had a chance to take a look? |
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
Okay, I think I have a grip on this now—nice contribution and great to see things like this wrapped up into BIDS Apps! To summarize, this project aims to compute beta-series correlations on BIDS-compliant data preprocessed using fMRIPrep. I have some general comments and then a laundry list of smaller-scale suggestions. For the really nit-picky stuff, I can make a PR if you don't mind. Also, bear in mind that I'm more neuroscientist than software developer per se, so apologies if any of these comments are way off the mark! General comments: I'm a little bit unsure about the Jupyter Notebook-style tutorial walkthrough in the "How to run" documentation. If I was planning to run this, I'd likely be running it from the Linux command line (maybe via a scheduler like Slurm on my server)—not invoking it via Python's subprocess. You jump through a bunch of hoops with Python just to download and modify the example data and only one cell of the tutorial actually runs This brings up the point that, if the preprocessed BIDS derivatives (e.g., in *_events.tsv) are fairly standard, should we expect Specific comments and questions:
|
This comment has been minimized.
This comment has been minimized.
jdkent
commented
Apr 1, 2019
|
Thank you so much for the in-depth review @snastase! I will be working on addressing these comments via issues/pull requests this week. |
This was referenced Apr 12, 2019
 whedon
assigned
snastase
whedon
assigned
snastase
May 6, 2019
This comment has been minimized.
This comment has been minimized.
danielskatz
commented
May 20, 2019
|
|
This comment has been minimized.
This comment has been minimized.
jdkent
commented
May 21, 2019
|
Hi @danielskatz, I am working through the comments still. I should have more time to dedicate to the project this week and the next. |
This comment has been minimized.
This comment has been minimized.
danielskatz
commented
May 21, 2019
|
Thanks for the update. |
whedon commentedMar 4, 2019
•
edited by snastase
Submitting author: @jdkent (James Kent)
Repository: https://github.com/HBClab/NiBetaSeries
Version: v0.2.3
Editor: @arokem
Reviewer: @snastase
Archive: Pending
Status
Status badge code:
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@snastase, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
The reviewer guidelines are available here: https://joss.theoj.org/about#reviewer_guidelines. Any questions/concerns please let @arokem know.
Review checklist for @snastase
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
paper.mdfile include a list of authors with their affiliations?