Join GitHub today
GitHub is home to over 40 million developers working together to host and review code, manage projects, and build software together.
Sign up[REVIEW]: Bedtoolsr: An R package for genomic data analysis and manipulation #1742
Comments
This comment has been minimized.
This comment has been minimized.
|
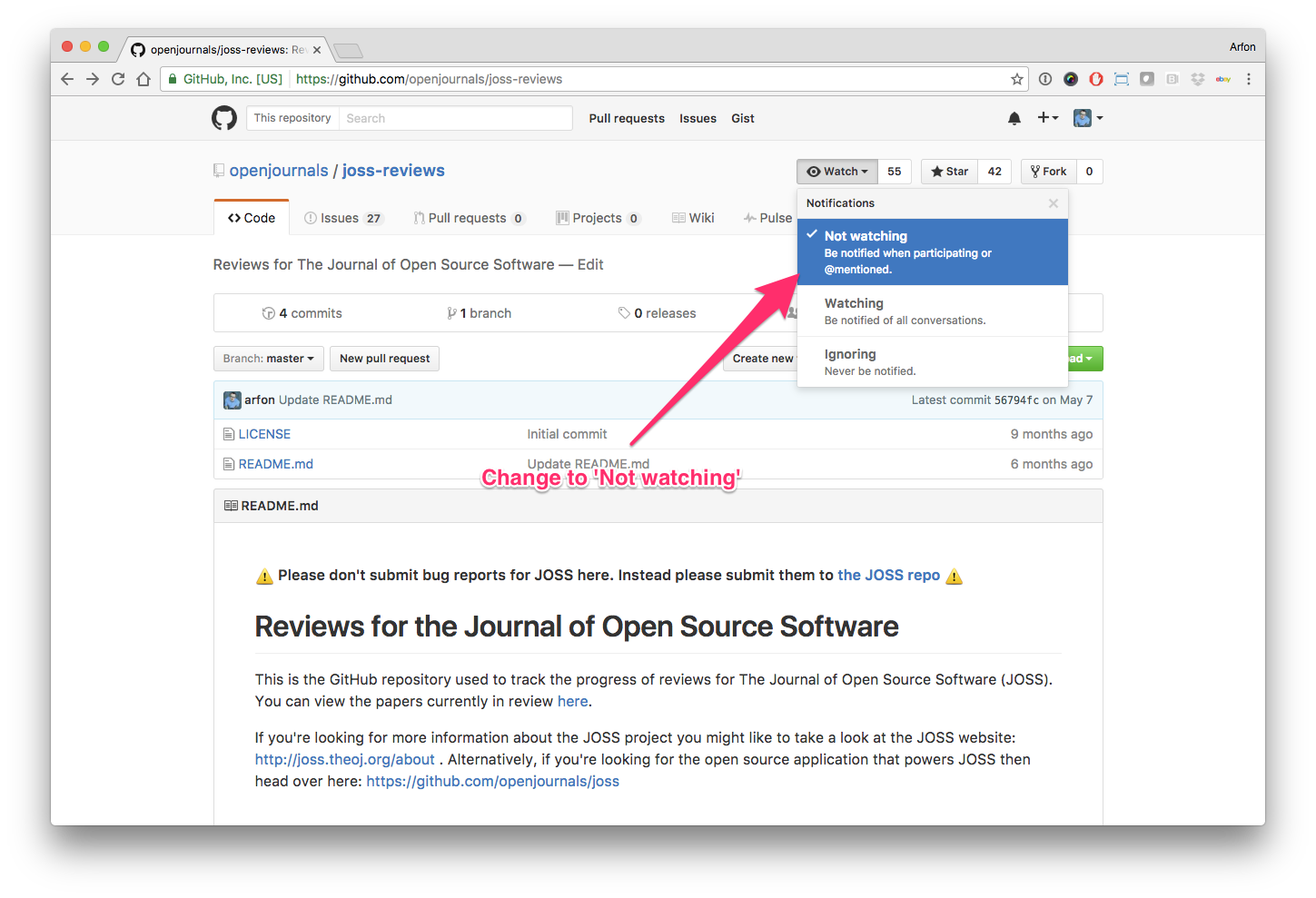
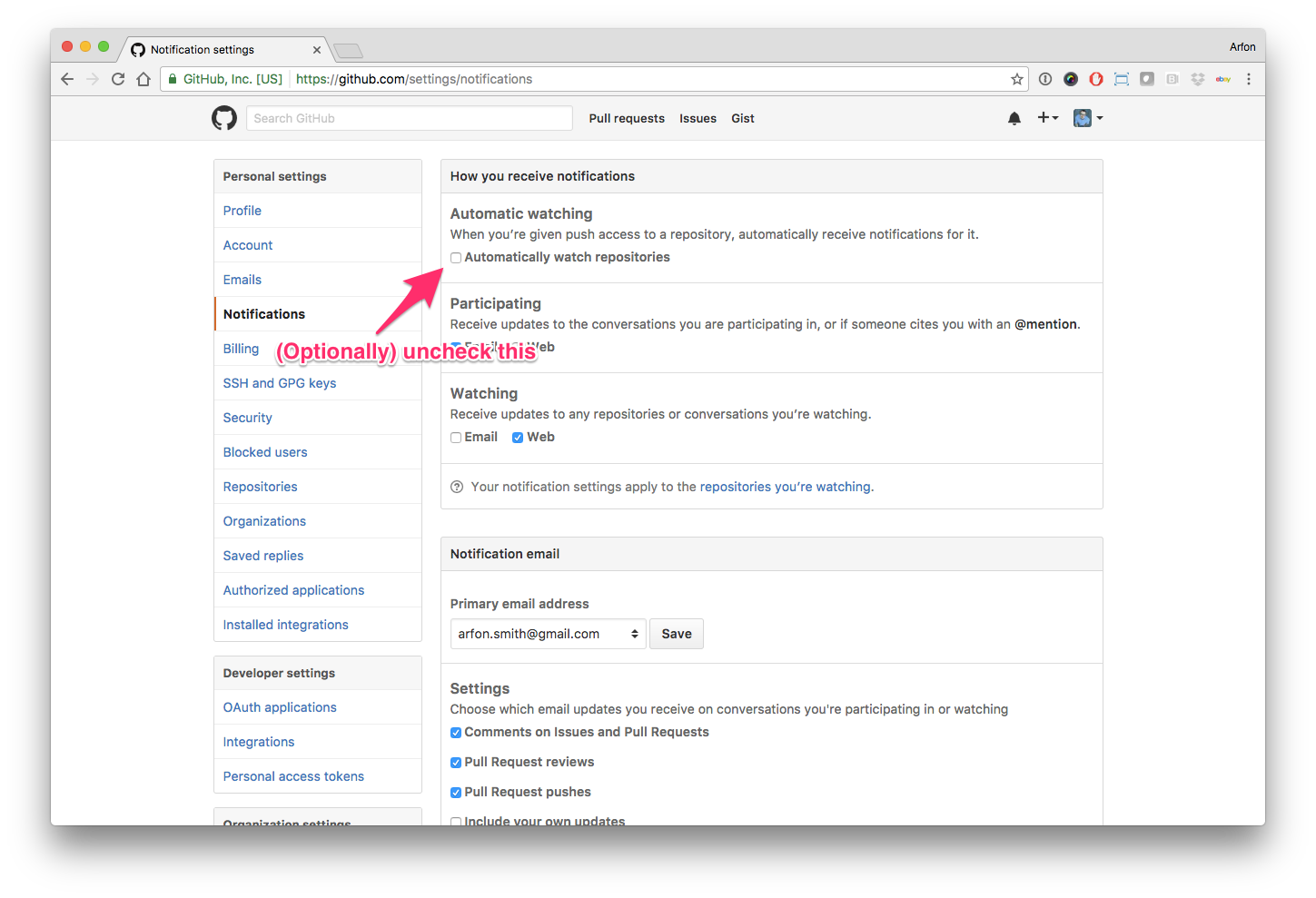
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @nhejazi , @mikmaksi it looks like you're currently assigned to review this paper If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews To fix this do the following two things:
For a list of things I can do to help you, just type: For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type: |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
Here is my review. Checklist above is also marked. Everything looks good except I could not find a License file. I have one minor suggestion for the installation instructions and to expand on the usage example. Thank you! General checks
Functionality
Documentation
Software paper
|
This comment has been minimized.
This comment has been minimized.
|
@dphansti et al., excellent work on this package, looks quite useful! After going through the review checklist, there's just a few items that require your attention before the few outstanding review items can be checked off. Please address the following when convenient:
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
Thank you so much for your quick and helpful reviews. We believe we have addressed all of the comments and issues. Mikmaksi’s comments
We followed reviewer nhejazi’s instructions below to properly handle licensing for the R package and for the repository.
Great suggestion. We have added the following text to README.md:
We have added a more complex and realistic example to the README.md file. nhejazi’s comments
We have added a LICENSE file, referenced by the DESCRIPTION file, with the copyright year and holders, a LICENSE.md with the full license information, and closed the issue. We greatly appreciate the reviewer’s detailed instructions in this regard.
We have added this short contribution guidelines section to README.md and closed the issue:
We have added DOIs for all journal article citations. |
This comment has been minimized.
This comment has been minimized.
|
Thanks for the quick work on this @dphansti --- all looks good to me! |
This comment has been minimized.
This comment has been minimized.
|
Look good to me as well. Thank you @dphansti |
This comment has been minimized.
This comment has been minimized.
|
@whedon generate pdf |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
@mikmaksi if all looks good can you complete the checklist? @dphansti this looks great. Can you just address these very minor comments on the proof? After that, can you make a Zenodo archive, ensuring the title and author list match the paper, and report the DOI here? Also please ensure the version listed here (v2.29.0) is correct. |
This comment has been minimized.
This comment has been minimized.
|
@mgymrek of course. I checked the last box that was not check (license) |
This comment has been minimized.
This comment has been minimized.
|
Thanks @mgymrek. I think we have addressed all of the issues.
Let me know if we need to do anything else. |
This comment has been minimized.
This comment has been minimized.
|
@whedon set v2.29.0-3 as version |
This comment has been minimized.
This comment has been minimized.
|
OK. v2.29.0-3 is the version. |
This comment has been minimized.
This comment has been minimized.
|
Thanks @dphansti. Can you just ensure the Zenodo archive and the paper have the same title? It looks like the Zenodo archive is titled "PhanstielLab/bedtoolsr: bedtoolsr v2.29.0-3". |
This comment has been minimized.
This comment has been minimized.
|
@mgymrek I edited the Zenodo archive title to match the paper title. Let me know if it still doesn't look right. Thanks. |
This comment has been minimized.
This comment has been minimized.
|
@whedon set 10.5281/zenodo.3509513 as archive |
This comment has been minimized.
This comment has been minimized.
|
OK. 10.5281/zenodo.3509513 is the archive. |
This comment has been minimized.
This comment has been minimized.
|
Thanks for the changes! @openjournals/joss-eics we're ready to accept this article! |
This comment has been minimized.
This comment has been minimized.
|
Great. Thank you all for your work on this. @mgymrek what happens next? Do we need to take any action? |
This comment has been minimized.
This comment has been minimized.
|
Pinging @openjournals/joss-eics again it looks like this might have been missed. |
This comment has been minimized.
This comment has been minimized.
|
@whedon generate pdf |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
please merge PhanstielLab/bedtoolsr#4 |
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
@danielskatz Both have been merged. Thanks. |
This comment has been minimized.
This comment has been minimized.
|
@whedon generate pdf |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
@whedon accept |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
Check final proof If the paper PDF and Crossref deposit XML look good in openjournals/joss-papers#1157, then you can now move forward with accepting the submission by compiling again with the flag |
This comment has been minimized.
This comment has been minimized.
|
@whedon accept deposit=true |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
|
This comment has been minimized.
This comment has been minimized.
|
Here's what you must now do:
Any issues? notify your editorial technical team... |
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
If you would like to include a link to your paper from your README use the following code snippets: This is how it will look in your documentation: We need your help! Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
|
Submitting author: @dphansti (Douglas Phanstiel)
Repository: https://github.com/PhanstielLab/bedtoolsr.git
Version: v2.29.0-3
Editor: @mgymrek
Reviewer: @nhejazi , @mikmaksi
Archive: 10.5281/zenodo.3509513
Status
Status badge code:
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@nhejazi & @mikmaksi , please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @mgymrek know.
Review checklist for @nhejazi
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
Review checklist for @mikmaksi
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper