Join GitHub today
GitHub is home to over 50 million developers working together to host and review code, manage projects, and build software together.
Sign up[REVIEW]: The pdb2sql Python Package: Parsing, Manipulation and Analysis of PDB Files Using SQL Queries #2077
Comments
|
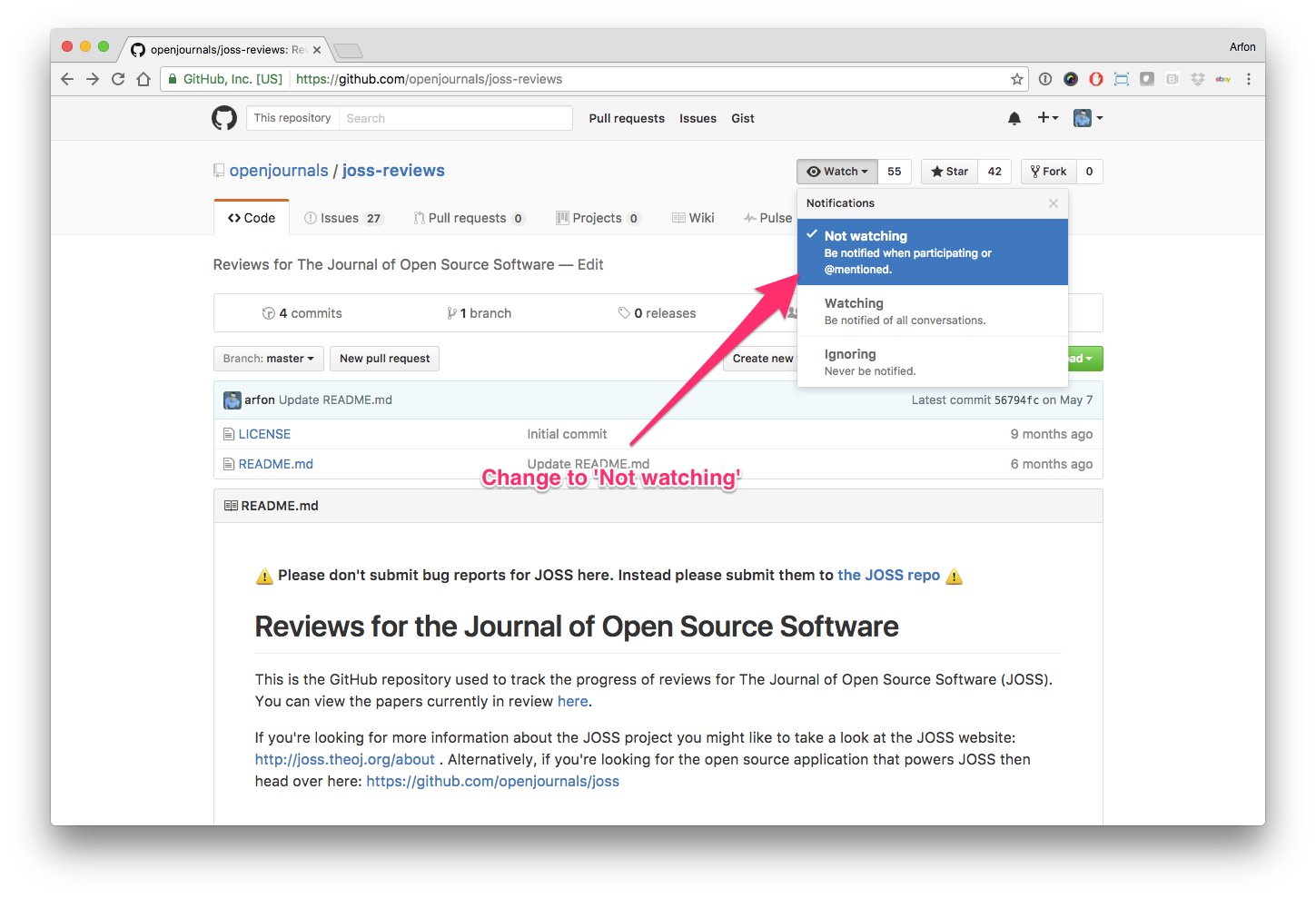
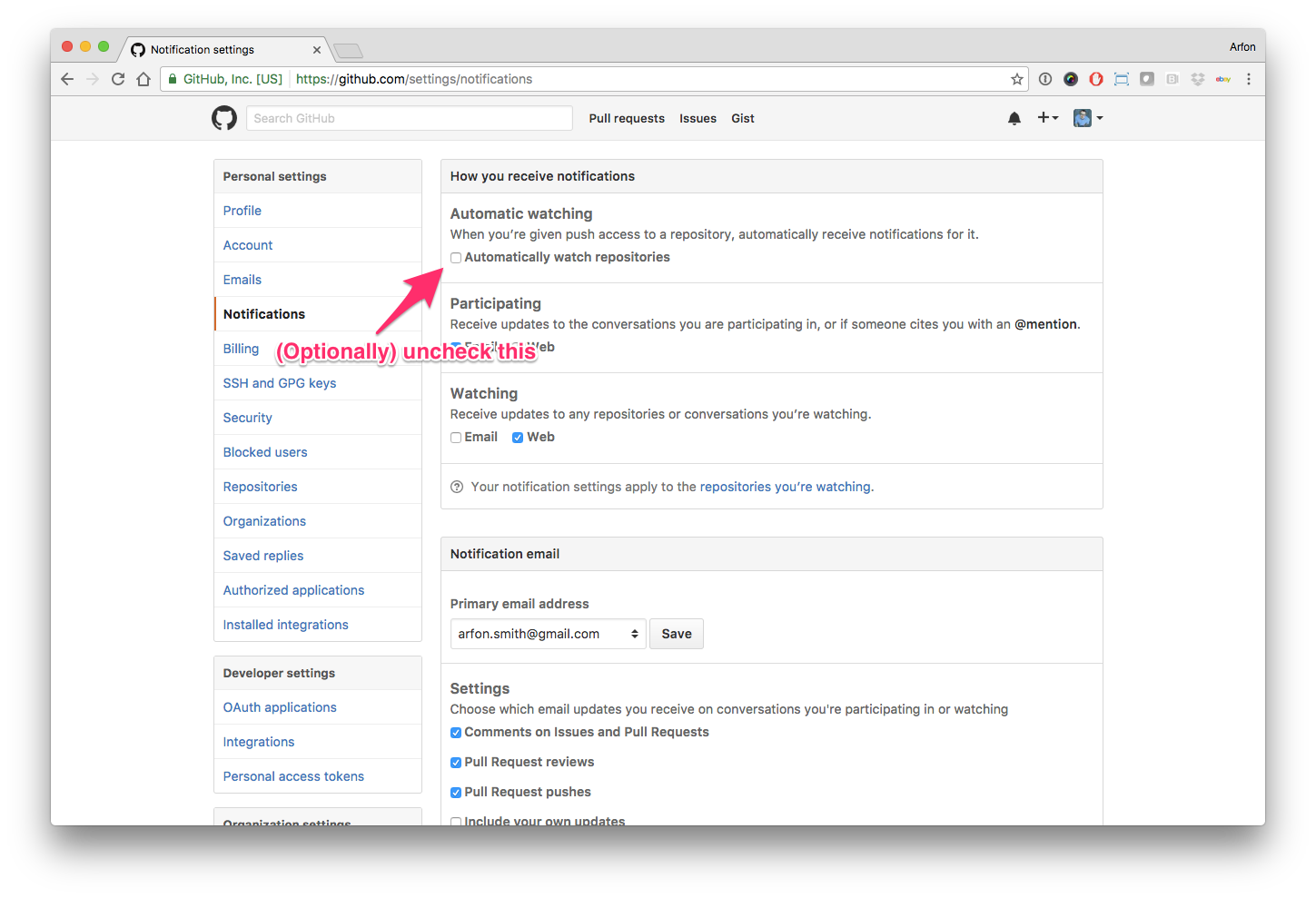
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @i-mtz, @JoaoRodrigues it looks like you're currently assigned to review this paper If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews To fix this do the following two things:
For a list of things I can do to help you, just type: For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type: |
|
|
@whedon generate pdf |
|
@JoaoRodrigues , @i-mtz, any update in the revision of this paper? thanks! |
|
Hi @lpantano , sorry for the late reply but I was traveling+sick and away from my laptop for quite some time. Apologies to the authors for the delay. |
|
Hey @NicoRenaud The tool looks good but there are a few rough edges I'd polish before publishing it. I left a few issues on your repo that should be easy to fix. Here are a few more minor questions/issues I found when going over the paper and the code:
Finally, I would send the text of the paper through Grammarly (its free version works well enough) to fix some incorrect grammar here and there and make it read more clearly. Overall, the tool does what it says it does and it's pretty quick too. As I said before, just some rough edges that need polishing! Good job @NicoRenaud and @CunliangGeng ! |
|
Dear @JoaoRodrigues, thanks for reviewing our code and for your comments and suggestions. We've addressed them all, merged the changes in the master and updated the paper accordingly. As a quick summary :
|
|
Dear authors and reviewers We wanted to notify you that in light of the current COVID-19 pandemic, JOSS has decided to suspend submission of new manuscripts and to handle existing manuscripts (such as this one) on a "best efforts basis". We understand that you may need to attend to more pressing issues than completing a review or updating a repository in response to a review. If this is the case, a quick note indicating that you need to put a "pause" on your involvement with a review would be appreciated but is not required. Thanks in advance for your understanding. Arfon Smith, Editor in Chief, on behalf of the JOSS editorial team. |
|
Hey @i-mtz, I hope all is good. Do you have an update for this tool? @JoaoRodrigues, were all your comments resolved? Thanks |
|
Hi @lpantano , I haven't had time to go over the changes in detail yet, but the message from @NicoRenaud seems to address most of my comments. I am a bit swamped today, but this is on my to-do list for this week. Is that OK? Sorry for the delay in reviewing |
|
All good, thank you! |
|
|
|
Hi @arfon @lpantano @NicoRenaud @CunliangGeng , this completely fell off my radar, I'm terribly sorry. Will reply as soon as possible! |
|
@whedon generate pdf |
|
Thank you for addressing my points! I'd only add somewhere in the documentation page a mention to the |
|
Sorry, I need to get a new reviewer since one of them couldn't make it under the current COVID-19 situation. @NicoRenaud , if you have a reviewer in mind, let me know. I will try to find another as soon as possible. |
|
Not to overstep my boundaries as a reviewer, but if I can suggest people: @joaomcteixeira or @jgreener64. Both very skilled structural bioinformaticians and very active on Github. |
|
@arfon we updated the instruction to manually run the test as pointed out by @joaomcteixeira. We could also follow his suggestion to change the exec path but I don't really have the time to do that at the moment. So from our side we are done I think |
|
@arfon just to let you know that @joaomcteixeira has implemented his own suggestions for the test so we are officially done with the review on our side ! Thanks again @joaomcteixeira !! |
|
Perfect! @NicoRenaud can you confirm the latest version is 0.2.2? if that is right, can you work on a zenodo archive of this version and having the title and authors matching what is shown in the paper? Thanks! |
|
@whedon check references |
|
|
@whedon generate pdf |
Thanks @lpantano, the version now is 0.4.0, and zenodo archive has been updated with your suggestions, see https://doi.org/10.5281/zenodo.3860329. |
|
@whedon set version as 0.4.0 |
|
I'm sorry human, I don't understand that. You can see what commands I support by typing: |
|
@whedon set 0.4.0 as version |
|
OK. 0.4.0 is the version. |
|
@whedon set 10.5281/zenodo.3860329 as archive |
|
OK. 10.5281/zenodo.3860329 is the archive. |
|
@whedon accept |
|
|
|
Check final proof If the paper PDF and Crossref deposit XML look good in openjournals/joss-papers#1460, then you can now move forward with accepting the submission by compiling again with the flag |
|
|
|
@danielskatz Many thanks for the changes, it's merged. |
|
@whedon accept deposit=true |
|
|
|
|
Here's what you must now do:
Any issues? notify your editorial technical team... |
|
Thanks to @i-mtz, @JoaoRodrigues, @joaomcteixeira for reviewing, and to @lpantano for editing! And congratulations to @NicoRenaud and co-author!! |
|
If you would like to include a link to your paper from your README use the following code snippets: This is how it will look in your documentation: We need your help! Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
|
Submitting author: @NicoRenaud (Nicolas Renaud)
Repository: https://github.com/DeepRank/pdb2sql
Version: 0.4.0
Editor: @lpantano
Reviewers: @i-mtz, @JoaoRodrigues, @joaomcteixeira
Archive: 10.5281/zenodo.3860329
Status
Status badge code:
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@i-mtz & @JoaoRodrigues & @joaomcteixeira, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @lpantano know.
Review checklist for @joaomcteixeira
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
Review checklist for @i-mtz
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
Review checklist for @JoaoRodrigues
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper