Join GitHub today
GitHub is home to over 50 million developers working together to host and review code, manage projects, and build software together.
Sign upDifferent results between MetFrag CLI and Web #39
Comments
|
Hi, I didn't yet get across to reproduce, but there was a recent report in #38 |
|
I don't think it's a mass rounding problem since mass values of pyrimidine and pyrazine are identical. |
|
This could be due to you using a different version of HMDB … the exact database used in MetFragWeb (in the screenshot you show) is available from here: https://zenodo.org/record/3375500
and should be used via the localCSV option with the CLI (rather than an SDF downloaded from HMDB). Can you confirm that this file gives you the same candidates as your web query?
|
|
Re mass rounding: this is not a CLI / Web version problem but seems specific to the local copy of PubChem … which contains apparently truncated masses (3 dp)
|
With "localCSV" option and the above CSV file, I could have two candidates as the same as web query. By the way, when I tried "localSDF" and SDF file (same mass query), I got some errors and could not obtaine the result. |
|
We also had issues with the HMDB SDF, hence we created and validated the CSV file. It's easier to add extra functionality to the CSV than the SDF. However, these local CSVs are a temporary solution and will eventually be integrated into a database so that you do not have to specify external files. This functionality will be released when ready but is not yet available. |
Hello,
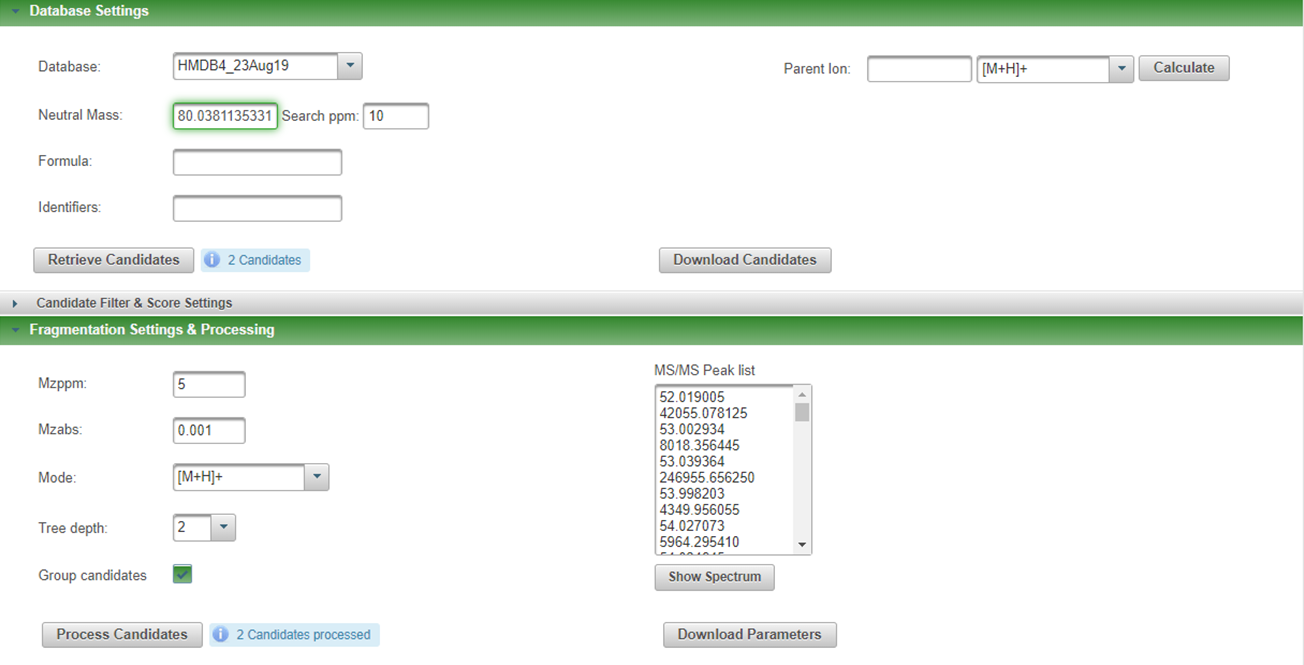
I ran MetFrag CLI and Web for a feature of neutral mass = 80.038114 against HMDB4, but got different results. CLI gave only pyrimidine as a candidate, but Web gave pyrimidine and pyrazine. Please let me know how I can get two candidates from CLI. For CLI, SDF file of HMDB4 was downloaded and used locally. Parameters are as follows,
PeakListPath = ms2.txt
LocalDatabasePath = hmdb.sdf
NeutralPrecursorMass = 80.038114
DatabaseSearchRelativeMassDeviation = 10
FragmentPeakMatchRelativeMassDeviation = 10
IsPositiveIonMode = True
PrecursorIonMode = 1
MetFragScoreTypes = FragmenterScore
MetFragScoreWeights = 1.0
MetFragCandidateWriter = CSV
SampleName = metfrag_result
ResultsPath = .
MaximumTreeDepth = 2
MetFragPreProcessingCandidateFilter = UnconnectedCompoundFilter
MetFragPostProcessingCandidateFilter = InChIKeyFilter
Thanks.
Screenshot of MetFragWeb
ms2.txt