Join GitHub today
GitHub is home to over 28 million developers working together to host and review code, manage projects, and build software together.
Sign upBuild error due to "Error in fromJSON(data) : no data to parse" #209
Comments
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
schymane
Oct 18, 2018
Member
|
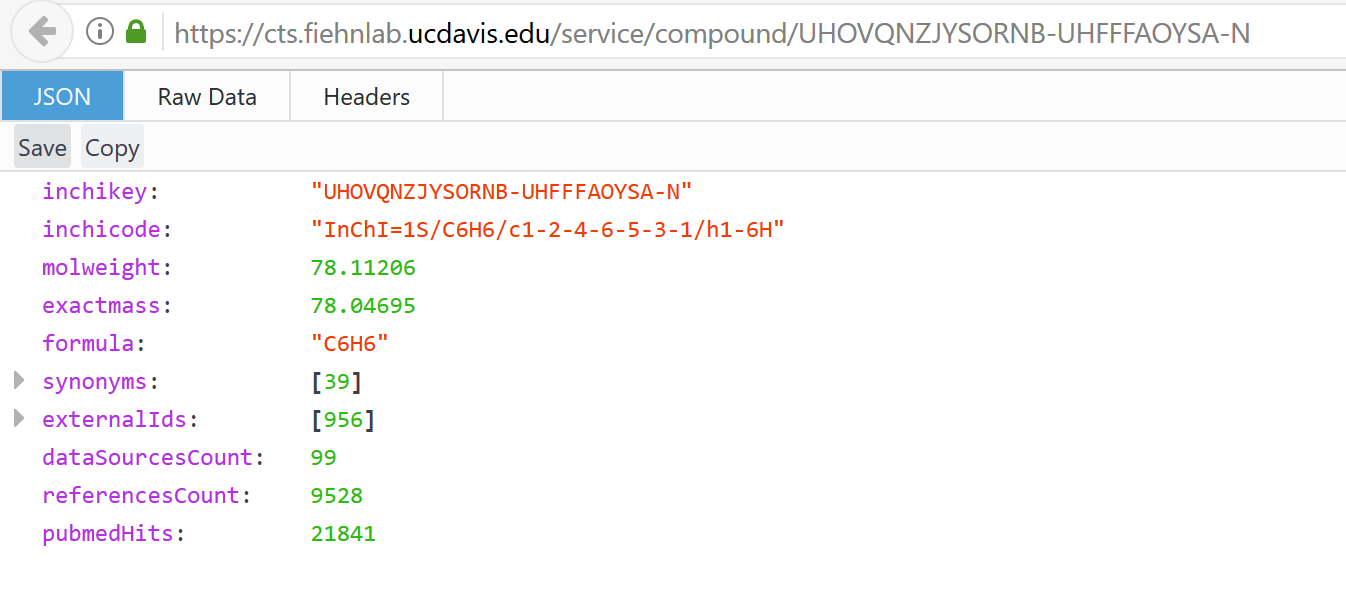
CTS seems to be working but looks different … have they changed their services?
https://cts.fiehnlab.ucdavis.edu/services
|
added a commit
that referenced
this issue
Oct 18, 2018
 schymane
closed this
in
#210
schymane
closed this
in
#210
Oct 18, 2018
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
schymane
Oct 18, 2018
Member
I just reinstalled but have the same error:
getCtsRecord("UHOVQNZJYSORNB-UHFFFAOYSA-N")
Error in fromJSON(data) : no data to parse
|
I just reinstalled but have the same error:
|
 schymane
reopened this
schymane
reopened this
Oct 18, 2018
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
schymane
Oct 18, 2018
Member
But the URL created in the code should work:
SessionInfo()
R version 3.4.2 (2017-09-28)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows >= 8 x64 (build 9200)
Matrix products: default
locale:
[1] LC_COLLATE=English_Australia.1252 LC_CTYPE=English_Australia.1252 LC_MONETARY=English_Australia.1252
[4] LC_NUMERIC=C LC_TIME=English_Australia.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RMassBank_2.9.1 Rcpp_0.12.19 devtools_1.13.3
loaded via a namespace (and not attached):
[1] BiocInstaller_1.26.1 compiler_3.4.2 git2r_0.19.0 plyr_1.8.4 rcdklibs_2.2.1
[6] bitops_1.0-6 iterators_1.0.10 ProtGenerics_1.8.0 tools_3.4.2 zlibbioc_1.22.0
[11] MALDIquant_1.16.4 digest_0.6.18 memoise_1.1.0 tibble_1.3.4 preprocessCore_1.38.1
[16] gtable_0.2.0 lattice_0.20-35 png_0.1-7 rlang_0.1.2 foreach_1.4.3
[21] curl_3.2 yaml_2.2.0 parallel_3.4.2 rJava_0.9-10 withr_2.0.0
[26] httr_1.3.1 IRanges_2.10.5 S4Vectors_0.14.7 stats4_3.4.2 grid_3.4.2
[31] Biobase_2.36.2 impute_1.50.1 R6_2.2.2 rcdk_3.4.9 XML_3.98-1.16
[36] BiocParallel_1.10.1 limma_3.32.10 ggplot2_2.2.1 mzR_2.10.0 itertools_0.1-3
[41] scales_0.5.0 pcaMethods_1.68.0 codetools_0.2-15 BiocGenerics_0.22.1 fingerprint_3.5.7
[46] mzID_1.14.0 MSnbase_2.2.0 colorspace_1.3-2 affy_1.54.0 RCurl_1.95-4.11
[51] doParallel_1.0.11 lazyeval_0.2.0 munsell_0.4.3 rjson_0.2.20 vsn_3.44.0
[56] affyio_1.46.0
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
tsufz
Oct 18, 2018
Member
Same for me:
library(RMassBank)
data <- getCtsRecord("UHOVQNZJYSORNB-UHFFFAOYSA-N")
Error in fromJSON(data) : no data to parse
sessionInfo()
R version 3.5.1 (2018-07-02)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows >= 8 x64 (build 9200)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.1252 LC_CTYPE=English_United Kingdom.1252 LC_MONETARY=English_United Kingdom.1252
[4] LC_NUMERIC=C LC_TIME=English_United Kingdom.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RMassBank_2.9.1 Rcpp_0.12.16
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 purrr_0.2.5 rJava_0.9-10 lattice_0.20-35 colorspace_1.3-2
[6] stats4_3.5.1 yaml_2.2.0 fingerprint_3.5.7 vsn_3.48.1 XML_3.98-1.16
[11] rlang_0.2.2 pillar_1.3.0 glue_1.3.0 MSnbase_2.6.4 mzR_2.14.0
[16] BiocParallel_1.14.2 affy_1.58.0 BiocGenerics_0.26.0 bindrcpp_0.2.2 affyio_1.50.0
[21] foreach_1.4.4 bindr_0.1.1 plyr_1.8.4 mzID_1.18.0 ProtGenerics_1.12.0
[26] zlibbioc_1.26.0 munsell_0.5.0 pcaMethods_1.72.0 gtable_0.2.0 rcdklibs_2.0
[31] codetools_0.2-15 Biobase_2.40.0 IRanges_2.14.12 doParallel_1.0.14 BiocInstaller_1.30.0
[36] parallel_3.5.1 itertools_0.1-3 preprocessCore_1.42.0 scales_1.0.0 limma_3.36.5
[41] rcdk_3.4.7.1 S4Vectors_0.18.3 impute_1.54.0 rjson_0.2.20 ggplot2_3.0.0
[46] png_0.1-7 digest_0.6.18 dplyr_0.7.7 grid_3.5.1 tools_3.5.1
[51] bitops_1.0-6 magrittr_1.5 lazyeval_0.2.1 RCurl_1.95-4.11 tibble_1.4.2
[56] crayon_1.3.4 pkgconfig_2.0.2 MASS_7.3-51 assertthat_0.2.0 rstudioapi_0.8
[61] iterators_1.0.10 R6_2.3.0 MALDIquant_1.18 compiler_3.5.1
|
Same for me:
Matrix products: default locale: attached base packages: other attached packages: loaded via a namespace (and not attached): |
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
tsufz
Oct 18, 2018
Member
I just started a conversation with Duncan, the maintainer of RCurl, but this stucked somehow. We checked some things, but it did not change. I dunno if something happens with RCurl... I wrote a workaround using httr:
get_cactus <- function(identifier,representation){
ret <- tryCatch(GET(paste("https://cactus.nci.nih.gov/chemical/structure/",
URLencode(identifier), "/", representation, sep = "")),
error = function(e) NA)
if (is.na(ret))
return(NA)
if (ret[2] == 404)
return(NA)
ret <- content(ret)
return(unlist(strsplit(ret, "\n")))
}
|
I just started a conversation with Duncan, the maintainer of RCurl, but this stucked somehow. We checked some things, but it did not change. I dunno if something happens with RCurl... I wrote a workaround using get_cactus <- function(identifier,representation){ } |
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
schymane
Oct 18, 2018
Member
|
I don’t get it … is this a load library problem? Do we have to change something in DESCRIPTION or somewhere else?
If I explicitly call library(RCurl) and library(rjson) it works and if not it doesn’t.
Here’s things working fine from “abgespekte” getCtsRecord code:
https://github.com/MassBank/RMassBank/blob/master/R/webAccess.R#L187
library(RCurl)
data <- getURL("https://cts.fiehnlab.ucdavis.edu/service/compound/UHOVQNZJYSORNB-UHFFFAOYSA-N")
library(rjson)
r <- fromJSON(data)
Restarting R session...
library(RMassBank)
Warning messages:
1: In fun(libname, pkgname) :
mzR has been built against a different Rcpp version (0.12.10)
than is installed on your system (0.12.19). This might lead to errors
when loading mzR. If you encounter such issues, please send a report,
including the output of sessionInfo() to the Bioc support forum at
https://support.bioconductor.org/. For details see also
https://github.com/sneumann/mzR/wiki/mzR-Rcpp-compiler-linker-issue.
2: package ‘Rcpp’ was built under R version 3.4.4
getCtsKey("UHOVQNZJYSORNB-UHFFFAOYSA-N")
[1] "UHOVQNZJYSORNB-UHFFFAOYSA-N"
getCtsRecord("UHOVQNZJYSORNB-UHFFFAOYSA-N")
Error in fromJSON(data) : no data to parse
Error in getURL("https://cts.fiehnlab.ucdavis.edu/service/compound/UHOVQNZJYSORNB-UHFFFAOYSA-N") :
could not find function "getURL"
library(RCurl)
Loading required package: bitops
Warning message:
package ‘RCurl’ was built under R version 3.4.4
data <- getURL("https://cts.fiehnlab.ucdavis.edu/service/compound/UHOVQNZJYSORNB-UHFFFAOYSA-N")
r <- fromJSON(data)
Error in fromJSON(data) : could not find function "fromJSON"
library(rjson)
Warning message:
package ‘rjson’ was built under R version 3.4.4
… r <- fromJSON(data)
|
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
tsufz
Oct 18, 2018
Member
In a pure R command line R --vanilla, I get the following:
library(RCurl)
getURL("http://cts.fiehnlab.ucdavis.edu/service/compound/UHOVQNZJYSORNB-UHFF
[1] ""
If I use the https URL, all seems to be fine:
getURL("https://cts.fiehnlab.ucdavis.edu/service/compound/UHOVQNZJYSORNB-UHFF
[1] ....
Here, the change to https may resolve the problem. The problem with CACTUS still persists:
getURL("https://cactus.nci.nih.gov/chemical/structure/LFTLOKWAGJYHHR-UHFFFAO$
Error in function (type, msg, asError = TRUE) :
Unknown SSL protocol error in connection to cactus.nci.nih.gov:443
|
In a pure R command line
If I use the https URL, all seems to be fine: Here, the change to https may resolve the problem. The problem with CACTUS still persists:
|
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
schymane
Oct 18, 2018
Member
Indeed ... this works:
https://cactus.nci.nih.gov/chemical/structure/UHOVQNZJYSORNB-UHFFFAOYSA-N/smiles
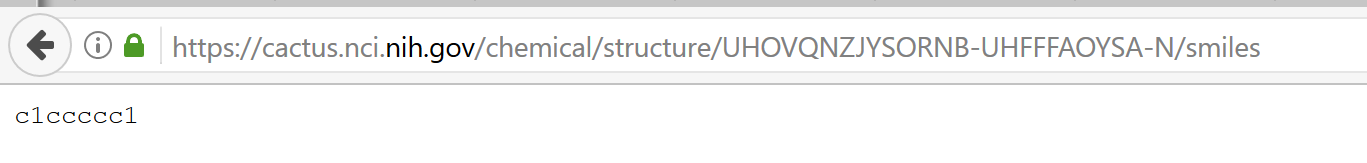
but not in R:
getURL("https://cactus.nci.nih.gov/chemical/structure/UHOVQNZJYSORNB-UHFFFAOYSA-N/smiles")
Error in function (type, msg, asError = TRUE) :
Unknown SSL protocol error in connection to cactus.nci.nih.gov:443
|
Indeed ... this works: but not in R:
|
This comment has been minimized.
Show comment
Hide comment
This comment has been minimized.
sneumann
Oct 22, 2018
Member
The issue started here was in particular on the CTS issue. Unfortunately
my patch #210 (merged by Emma, thanks!) fixed only one occurrence
of the baseURL , so I committed 1ce4d16 to fix the rest.
We still need someone (@meowcat ?) to push to BioC.
The Unknown SSL protocol error is a separate Windows / RCurl issue,
please use #208 to discuss that. Yours, Steffen
|
The issue started here was in particular on the CTS issue. Unfortunately We still need someone (@meowcat ?) to push to BioC. The |
sneumann commentedOct 18, 2018
Hi, on BioC we currently get a build error on all platforms due to
resulting in
Error in fromJSON(data) : no data to parse.=> Is that RMB or an issue on CTS side ?
Yours, Steffen