Join GitHub today
GitHub is home to over 31 million developers working together to host and review code, manage projects, and build software together.
Sign upPatch/get mass #547
Conversation
johnmay
added some commits
Feb 18, 2019
This comment has been minimized.
This comment has been minimized.
|
@schymane... hard to find you. Will also ping in CDK-R/cdkr#65 |
egonw
requested changes
Feb 18, 2019
|
Thanks, I like the API. I also like your suggestion of a |
| * a distribution of isotopes by splitting the set of isotopes in two, | ||
| * the one under consideration (specified by 'idx') and the remaining to be | ||
| * considered ('>idx'). The inflection point is calculate as 'k' | ||
| * &le 'count' isotopes added. If there are remaining isotopes the method |
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 3, 2019
|
So, it seems you have built in 4 cases and we had 5 in the latest changes @rajarshi and I worked on in rcdk ... the missing one is the mass integer (maybe worth adding for our nominal mass friends?). Our test cases don't overlap which is making the terminology match confusing. https://github.com/CDK-R/cdkr/blob/massfuncs/rcdk/inst/unitTests/runit.mass.R I cannot figure out if these overlap completely with the different terms and the unfamiliar masses ... I suspect one option doesn't - and I think once we have all the options we need, we should come to an agreement on names. It's not clear for mass spec people which one is monoisotopic mass in our rcdk terms, it is for yours, but it's not clear if and when isotopes are ignored and this is very confusing for end users. Are you able to implement a couple of my trickier test cases? I chose them to check the implicit/explicit H handling (a nasty case for deuterium, especially deuterium exchange) and the isotope handling as well. |
This comment has been minimized.
This comment has been minimized.
|
I kind of think MonoIsotopic should also me modified such that explicitly specified isotopes are considered (same as MolWeight) currently it just throws them away.. In terms of naming, happy to modify and drop the Weight from AverageWeight as needed but "total.exact" and "total.natural" are non starters for me, works in R but not in Java - we can't have the "dots" and TotalExact TotalNautral meh. As you say "MolWeight" is too ubiquitous, and we have to think about the community in general rather just MS so I would keep that, it's currently the default which I think makes sense.
Not really a fan of that duplication, think it makes it more confusing. |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 3, 2019
I agree that it's "wrong" but agree we have to leave it in for backwards compatibility.
I confess I was severely undercaffeinated this morning and found those definitions in my emails after giving my feedback, sorry :-/ and thanks for pasting again
OK this is good, behaviour is "normal" unless we specify, and the name is what most would consider logical.
As you said, it's needed for backwards compatibility and having the "weight" in there will connect it back to MolWeight. I would not know a chemically correct term for this offhand since conceptually it's not really logical. Suggest to add "(this option is retained for backwards compatibility)" or similar to the documentation, so purists that have an issue with a rather illogical calculation know the reasoning?
This is an absolute killer for mass spec if this is the only exact mass option and we'd need an additional function that gives us the exact mass for specified isotopes. So that's the case that was missing. If we go strictly by definition (which is from the IUPAC Gold Book): Then we will need two cases. CDK's MonoIsotopic option should match the IUPAC definition (and does if your major isotope the principal by abundance - maybe adjust the definitions to be as close to the IUPAC as possible?), then we would need another option ExactMass (or similar) to return the exact mass of the exact isotopic combination specified. , i.e
Agree with the need for this and like the name. It says what it is.
See above - we should match the IUPAC definition.
Sorry I was meaning we should match terms in terms of "getting our words consistent" and ignoring the obvious styling differences between the languages. Whether we use caps or dots to do the separation doesn't really matter (and we'll need to keep the dots to keep the syntax in rcdk consistent) but I think most humans can match AvgWeight v avg.weight and ExactMass v exact.mass ;-) and this is what we need to get consistent.
Fine with me - I agree in this case that it could add too much confusion. I would strongly suggest to put in some of the examples I suggested, or similar, as they were selected to show the need for the two different styles of exact mass - and will also check the H behaviour. Or I can give it a go once we have all the options if you trust my copy-paste work ;-) |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 3, 2019
I spoke too soon, I find the definition a little unclear, suggest a minor wording change (if I understood it correctly...seems to match the enviPat output at least ... ):
For the record: |
This comment has been minimized.
This comment has been minimized.
|
Sorry miss-documented! I propose the following, we can have ugly names for the ones we don't want people to use:
|
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 3, 2019
I'm not yet convinced it does ... comments below ... and I've finally found the good overview:
I find MolWeight clearer than Average. Molecular weight is the term commonly used. Note at the above site, MolWeight (which is calls MolMass ;-) ) and Average mass are defined separately. Would much rather MolWeight for clarity.
I'm concerned here that you deviate from the IUPAC definition, because it specifies the exact isotopic state for this mass: I think this should be "Exact" (given your comment you don't want repetition).
OK for ugly name here but then please MolWeightIgnoringSpecified? Or rather IgnoringIsotopes?
See above I think this should be MonoIsotopic (it gives people the MonoIso mass irrespective of the input isotopes, which is exactly in line with the IUPAC definition; Exact will give MonoIso only if isotopes are unspecified) - there will be cases for mass spec where this is very useful
All good with this one still as far as I can tell
Looks like you hit the nail on the head ... Do you see a need for the Nominal Mass in addition? As far as I can tell it's just a simple operation from MonoIso and shouldn't need to be hard coded (and becoming slowly obsolete ...) |
This comment has been minimized.
This comment has been minimized.
|
Maybe there is something I'm missing but I need a bit more explanation on the difference between |
This comment has been minimized.
This comment has been minimized.
|
The crux of the issue is "What do you do when an isotope is unspecified?" If everything is specified The So if all were unspecified the exact mass is the same as mono isotopic? |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
As far as I can tell, MonoIsotopic and MostAbundant should be fixed, irrespective of the isotopes defined (because they have fixed definitions wrt which isotopes to use - or is it me misinterpreting the definition?), whereas Exact would give the exact mass for the exact isotopic state given in the input. Thus as I see it, Exact is the generic function and MonoIsotopic and MostAbundant are two, fixed "special cases". Exact should default to MonoIsotopic if isotopes are not defined. This is at least what I set up in my test cases for rcdk (but I did not have MostAbundant so those numbers are missing): |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
This should not be the case ...
Yes, if isotopes are unspecified, MonoIsotopic = Exact |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
...but of course here not every isotope is specified ... I think I misunderstood ... OK so we need a test case with every single individual isotope defined too. |
This comment has been minimized.
This comment has been minimized.
|
What do you mean by "fixed" do you mean should not change if the isotopic definitions change? Do you think the value should change if I gave it D2O vs H2O currently these will give different values for both If a user specifies absolutely everything the mass functions all return the same thing, the option is really what do you do when there is missing information. |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
Taking the definitions strictly, these values should not change irrespective of the isotopic definitions, because the terms MostAbundant and MonoIsotopic define the isotopes to use (as currently written). Hence Exact is the generic form (and should probably be the setting to encourage people to use if they want to get either the Exact or MonoIsotopic mass). MonoIsotopic is still in there for the "normal" cases and also for backwards compatibility. I think anything else will end up giving masses that aren't really "defined" with a concept behind them, although I am on the fence about MostAbundant ... I need to see the numbers side by side. It might make more sense if this shifts with the given isotopes but then you have to assume the others are "normal" if they are only partially defined? What makes most sense with your current code base?
I need to see numbers ...
We should add this as a test case. Should I expand yours, or will you do? |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
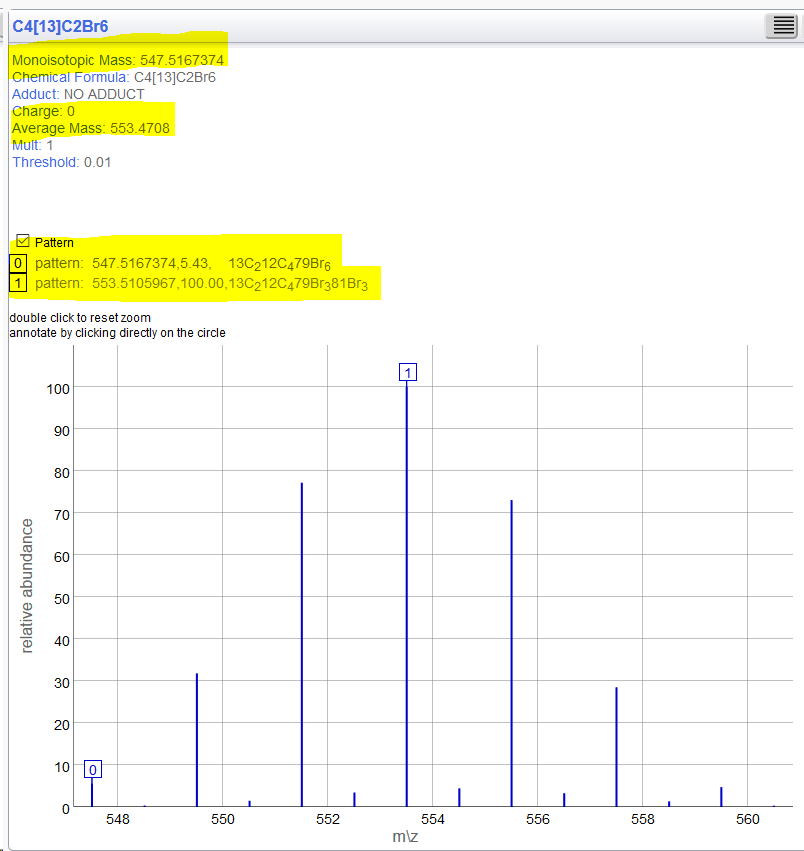
[12]C4[13]C2Br6 Exact = 547.5167 - in this picture also matches MonoIsotopic but this is not strictly in keeping with the definition(?). According to the IUPAC definition it should be 545.51? But if everywhere shifts this according to defined isotopes, we should be consistent. Then you just need to maintain backwards compatibility with the functionality that ignored isotopes with another setting? |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
I would still prefer for clarity to name it "Exact" mass - and maybe revert to your suggestion a few comments back with MonoIsotopicIgnoresIsotopes for the backwards compatible function? In most cases MonoIso = Exact up until now but times are changing with lots more isotopic experiments going on and we should encourage correct usage ...
|
This comment has been minimized.
This comment has been minimized.
|
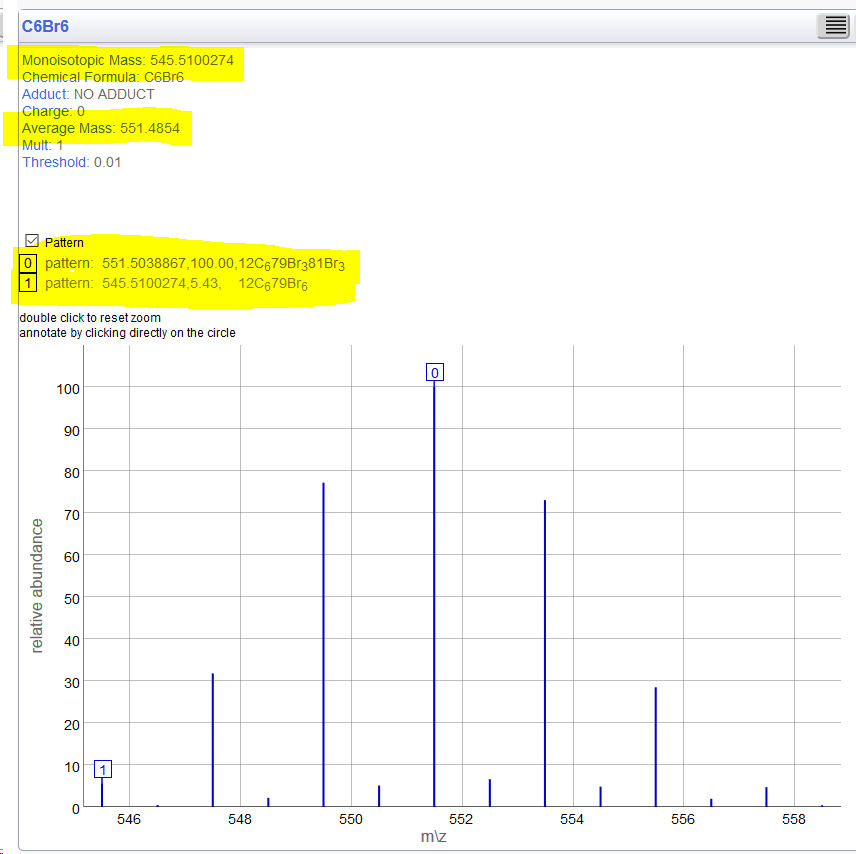
So I match those screenshots... @Test
public void C6Br6() {
assertMass("C6Br6", 551.485, MolWeight);
assertMass("C6Br6", 545.510, MonoIsotopic);
assertMass("C6Br6", 551.503, MostAbundant);
assertMass("[12]C4[13]C2Br6", 553.427, MolWeight);
assertMass("[12]C4[13]C2Br6", 547.516, MonoIsotopic);
assertMass("[12]C4[13]C2Br6", 553.510, MostAbundant);
}
|
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
Looks OK if you call it Exact and not MonoIsotopic?
Want more screenshots for your other examples? If so, pls copy paste me the formulas you want ;-)
|
This comment has been minimized.
This comment has been minimized.
I still prefer |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
It’s not the strict definition … a lot of German-made software call Molecular Formula the “Sum Formula” (Summenformel), which is now turning up in manuscripts – just because a term is used wrong, doesn’t mean we should propagate the error?
Maybe some other people can add their thoughts @rajarshi @egonw @meowcat @blosloos @lauperbe @hunter-moseley
We are debating the use of MonoIsotopic Mass (as in below) vs Exact Mass:
@test
public void C6Br6() {
assertMass("C6Br6", 551.485, MolWeight);
assertMass("C6Br6", 545.510, MonoIsotopic);
assertMass("C6Br6", 551.503, MostAbundant);
assertMass("[12]C4[13]C2Br6", 553.427, MolWeight);
assertMass("[12]C4[13]C2Br6", 547.516, MonoIsotopic);
assertMass("[12]C4[13]C2Br6", 553.510, MostAbundant);
}
Unfortunately an even number so we could get a deadlock ;)
https://en.wikipedia.org/wiki/Monoisotopic_mass
The monoisotopic mass is the sum of the masses of the atoms in a molecule using the unbound, ground-state, rest mass of the principal (most abundant) isotope for each element instead of the isotopic average mass.
https://en.wikipedia.org/wiki/Mass_(mass_spectrometry)#Exact_mass
The exact mass of an isotopic species (more appropriately, the calculated exact mass[8]<https://en.wikipedia.org/wiki/Mass_(mass_spectrometry)#cite_note-ODS-8>) is obtained by summing the masses of the individual isotopes of the molecule.
|
This comment has been minimized.
This comment has been minimized.
|
Just clarify something you would argue the strict definition of MonoIsotopic means even the MF with 13C should be 545.510? That is information provided is ignored. |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 4, 2019
|
If you interpret strictly, yes. Whether this is sensible or not is another matter … interested in what @hunter-moseley thinks on that one …
|
This comment has been minimized.
This comment has been minimized.
hunter-moseley
commented
Mar 5, 2019
|
The definitions of monoisotopic mass, exact mass, and most abundant mass
are based on an elemental molecular formula perspective.
When isotopes are defined explicitly in an isotope-resolved molecular
formula, then there is only one mass and these definitions have no meaning.
The problem appears to be how to interpret these three mass definitions
when you have a partially isotope-resolved molecular formula.
I think Emma is suggesting that the molecular formula is divided into two
parts: an isotope-resolved part and an element-resolved part.
The isotope-resolved part has only one mass, while the element-resolved
part has a different mass based on the three definitions.
Then these two parts are summed to get the mass based on the three
different definitions.
This sounds reasonable to me, but could get complicated if both
isotope-resolved and element-resolved parts are shared for the same element.
For example: "C4[13]C2Br6".
However, you could still separate the isotope-resolved part from the
element-resolved part and calculate accordingly.
That is my two-cents worth of interpretation.
Cheers,
Hunter
…On Mon, Mar 4, 2019 at 7:38 AM Emma Schymanski ***@***.***> wrote:
If you interpret strictly, yes. Whether this is sensible or not is another
matter … interested in what @hunter-moseley thinks on that one …
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#547 (comment)>, or mute the
thread
<https://github.com/notifications/unsubscribe-auth/AMj_hyLYtC2mVZnW0e7BfysGhOrKJjdgks5vTROsgaJpZM4bA6wv>
.
--
Hunter Moseley, Ph.D. -- Univ. of Kentucky
Associate Professor, Dept. of Molec. & Cell. Biochemistry / Markey Cancer
Center
/ Resource Center for Stable Isotope Resolved Metabolomics
Not just a scientist, but a fencer as well.
My foil is sharp, but my mind sharper still.
---------------------------------------------------------------
Email: hunter.moseley@uky.edu (work) hunter.moseley@gmail.com
(personal)
Phone: 859-218-2964 (office) 859-218-2965 (lab) 859-257-7715 (fax)
Web: http://bioinformatics.cesb.uky.edu/
Address: CC434 Roach Building, 800 Rose Street, Lexington, KY 40536-0093
|
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 5, 2019
Based on Hunter's response, I think we can stick to MonoIsotopic. |
This comment has been minimized.
This comment has been minimized.
|
OK, I'll remove the |
This comment has been minimized.
This comment has been minimized.
schymane
commented
Mar 5, 2019
|
As long as this won’t cause backwards compatibility issues?
I know from experience that a lot of our functions have built workarounds on the old (incorrect) behaviour and we are not the only ones … this could cause a cascade of errors if those functions are removed entirely ….
|
johnmay commentedFeb 18, 2019
Adds
getMass()functions to AtomContainerManipulator and MolecularFormulaManipulator unify the existing calculations. Existing methods are deprecated pointing to the new API.Some open questions, I used integers for the options but a new 'MassType' enum might be useful. I really don't like adding new class but in this case might make sense. Naming of the options is flexible, would be good to get Emma's thoughts on this (can't find her GitHub handle ATM).
The tests summarise it nicely: