GraphPass
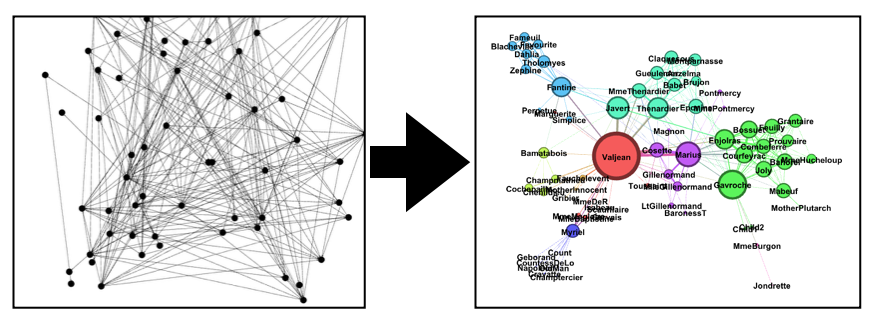
GraphPass is a utility to filter networks and provide a default visualization output for Gephi or SigmaJS. It prevents the infamous "borg cube" result when entering large files into Gephi, allowing you to work with ready-made network layouts.
Installation
Dependencies
This utility requires the C Igraph Library and a C compiler, such as gcc.
For Linux (Ubuntu):
Install the igraph dependencies:
sudo apt-get install git gcc libxml2-dev build-essential
wget http://igraph.org/nightly/get/c/igraph-0.7.1.tar.gz
tar -xvzf igraph-0.7.1.tar.gz
cd igraph-0.7.1
./configure
make
sudo make install
sudo ldconfig
You can test that gcc and igraph have been installed properly by using:
gcc -ligraph
If you get undefined reference to 'main' that means Linux is looking for
graphpass and you are ready to go.
If you get cannot find -ligraph then something went wrong with the install.
Check through the logs to see what failed to install.
If desired remove the igraph directory:
cd ..
rm -rf igraph-0.7.1
For MacOS:
Using brew, the following commands will install dependencies:
brew install gcc
brew install igraph
Type
brew info igraph
and verify that the path displayed there matches the default IGRAPH_PATH value provided in the Makefile. By default this is /usr/local/Cellar/igraph/0.7.1_6/ for MacOS.
Building
Go to your preferred install directory, for example:
cd /Users/{USERNAME}/
Clone the repository with:
git clone https://github.com/archivesunleashed/graphpass
Then
cd graphpass
make
Usage
Once compiled use the following command:
./graphpass {FLAGS}
The following flags are available:
--file {FILENAME}- sets the default filename. If not set, graphpass will use a default network in /assets.--dir {DIRECTORY}- the path to look for {FILENAME} by default this isassets/--output {OUTPUT}- the directory to send output files such as filtered graphs and data reporst.--percent {PERCENT}- a percentage to remove from the file. By default this is 0.0.--method {options}- a string of various methods through which to filter the graph.
These various methods are outlined below:
a: authorityb: betweennessd: simple degreee: eigenvectorh: hubi: in-degreeo: out-degreep: pagerankr: random
For example:
./graphpass --percent 10 --methods b --file links-for-gephi.graphml --output OUT/
Will remove 10% of the graph using betweenness as a cutting measure and lay the network out. It will find links-for-gephi.graphml file in /assets and output a new one to /OUT (titled links-for-gephi10Betweenness.graphml).
Optional arguments
--reportor-r: create an output report showing the impact of filtering on graph features.--no-saveor-n: does not save any filtered files (useful if you just want a report).--quickor-q: provides a "quickrun" for basic--gexfor-g: output as a .gexf (e.g. for SigmaJS inputs) instead of .graphml.
Troubleshooting
It is possible that you can get a "error while loading shared libraries" error in Linux. If so, try running sudo ldconfig to set the libraries path for your local installation of igraph.
License
Licensed under the Apache License, Version 2.0.
Acknowledgments
This work is primarily supported by the Andrew W. Mellon Foundation. Additional funding for the Toolkit has come from the U.S. National Science Foundation, Columbia University Library's Mellon-funded Web Archiving Incentive Award, the Natural Sciences and Engineering Research Council of Canada, the Social Sciences and Humanities Research Council of Canada, and the Ontario Ministry of Research and Innovation's Early Researcher Award program. Any opinions, findings, and conclusions or recommendations expressed are those of the researchers and do not necessarily reflect the views of the sponsors.
The author would also like to thank Drs. Ian Milligan & Jimmy Lin plus Nick Nuest and Samantha Fritz for their kind advice and support.