-
Be the first to like this
-
The AI Rush by Jean-Baptiste Dumont 1045326 views
-
AI and Machine Learning Demystified... by Carol Smith 3617894 views
-
10 facts about jobs in the future by Pew Research Cent... 660287 views
-
2017 holiday survey: An annual anal... by Deloitte United S... 1068247 views
-
Harry Surden - Artificial Intellige... by Harry Surden 621931 views
-
Inside Google's Numbers in 2017 by Rand Fishkin 1204920 views
FSUJena Environmental Cheminformatics to Identify Unknowns April 2019
Emma Schymanski
, PI, Environmental Cheminformatics at LCSBPublished on
The Environmental Cheminformatics group at the Luxembourg Centre for Systems Biomedicine focuses on the comprehensive identification of known and unknown chemicals in our environment to investigate their effects on health and disease. The environment and the chemicals to which we are exposed is incredibly complex, with over 125 million chemicals registered in the largest chemical registry and over 70,000 in household use alone. Detectable molecules in complex samples can now be captured using high resolution mass spectrometry (HRMS), which provides a “snapshot” of all chemicals present in a sample and allows for retrospective data analysis through digital archiving. However, scientists cannot yet identify the vast majority of the tens of thousands of features in each sample, leading to critical bottlenecks in identification and data interpretation. For instance, recent studies indicate a strong connection between the gut microbiome and Parkinson’s disease, yet over 60 % of significant metabolites in microbiome experiments are unknown. Unknown identification remains extremely time consuming and, in many cases, a matter of luck. Prioritizing efforts to find significant metabolites or potentially toxic substances responsible for observed effects is the key, which involves reconciling highly complex samples with expert knowledge and careful validation. This talk will cover European, US and worldwide community initiatives to help connect knowledge on chemistry and toxicity with environmental observations - from compound databases to spectral libraries and retrospective screening. It will touch on the challenges of standardized structure representations, data curation, deposition and communication between resources. Finally, it will show how interdisciplinary efforts and data sharing can facilitate research in metabolomics, exposomics and beyond. Kindly hosted by Prof. Christoph Steinbeck
FSUJena Environmental Cheminformatics to Identify Unknowns April 2019
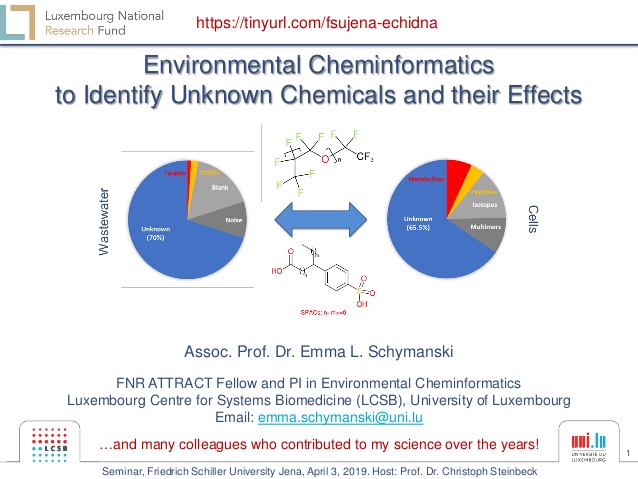
- 1. 1 Environmental Cheminformatics to Identify Unknown Chemicals and their Effects Assoc. Prof. Dr. Emma L. Schymanski FNR ATTRACT Fellow and PI in Environmental Cheminformatics Luxembourg Centre for Systems Biomedicine (LCSB), University of Luxembourg Email: emma.schymanski@uni.lu …and many colleagues who contributed to my science over the years! https://tinyurl.com/fsujena-echidna Seminar, Friedrich Schiller University Jena, April 3, 2019. Host: Prof. Dr. Christoph Steinbeck
- 2. 2 University of Luxembourg & LCSB o Uni Lu was founded in 2003 • Teenage years! o LSCB was founded in 2009 • …and is still pre-teenager • Young and very dynamic working environment!
- 3. 3 Environmental Cheminformatics and Biomedicine? Chemistry what? Medicine why? Biology how? Patient cohort Yeast MouseHuman Zebrafish Microbiome
- 4. 4 Environmental Cheminformatics … the Group S. Gene; https://en.wikipedia.org/wiki/File:Zwei_zigaretten.jpg; R. Singh; DOI:10.1186/s13321-017-0223-1; DOI: 10.1016/j.aca.2017.12.034 Sources:
- 5. 5 Our challenge? We still have many unknowns … (l) Data from Schymanski et al 2014, ES&T DOI: 10.1021/es4044374. (r) E. coli data provided by N. Zamboni, IMSB, ETH Zürich. Wastewater Cells
- 6. 6 1 10 100 1000 10000 100000 1 million 1 billion chemicals …. …. …. Our (Community) Challenge: Identifying Chemicals Data: Schymanski et al 2014, Environ. Sci. Technol. DOI: 10.1021/es4044374; Hollender et al 2017 DOI: 10.1021/acs.est.7b02184 Sample High resolution mass spectrometry
- 7. 7 1 10 100 1000 10000 100000 1 million 1 billion chemicals …. …. …. Our (Community) Challenge: Identifying Chemicals Data: Schymanski et al 2014, DOI: 10.1021/es4044374; https://www.slideshare.net/EmmaSchymanski/small-molecules-in-big-data-analytica-munich Sample High resolution mass spectrometry Chemicals AND connecting chemical knowledge
- 8. 8 Target, Suspect and Non-Target Screening KNOWNS SUSPECTS No Prior Knowledge HPLC separation and HR-MS/MS TARGET ANALYSIS SUSPECT SCREENING NON-TARGET SCREENING Targets found Suspects found Masses of interest (Molecular formula) DATABASE SEARCH STRUCTURE GENERATION Confirmation and quantification of compounds present Candidate selection (retention time, MS/MS, calculated properties) Sampling extraction (SPE) HPLC separation HR-MS/MS Time, Effort & Number of Compounds…. SUSPECTS SPECTRUM SEARCH Spectral match
- 9. 9 Target Analysis: Status Quo (>364 targets) Schymanski et al. (2014), ES&T, 48: 1811-1818. DOI: 10.1021/es4044374 Target List HPLC separation and HR-MS/MS TARGET ANALYSIS Targets found Confirmation and quantification of compounds present Sampling extraction (SPE) HPLC separation HR-MS/MS TPs!
- 10. 10 Target Analysis: Status Quo (>364 targets) Schymanski et al. (2014), ES&T, 48: 1811-1818. DOI: 10.1021/es4044374 Target List HPLC separation and HR-MS/MS TARGET ANALYSIS Targets found Confirmation and quantification of compounds present Sampling extraction (SPE) HPLC separation HR-MS/MS
- 11. 11 Suspect Screening: Different Approaches Target List Suspect List HPLC separation and HR-MS/MS TARGET ANALYSIS SUSPECT SCREENING Targets found Suspects found Confirmation and quantification of compounds present Candidate selection (retention time, MS/MS, calculated properties) Sampling extraction (SPE) HPLC separation HR-MS/MS o Search in mass spectral libraries o Screen for predicted transformation products of known parent compounds o Look for “well known” substances without reference standards o Screen for known homologue series
- 12. 12 Searching Mass Spectral Libraries o … which one? Peisl, Schymanski & Wilmes, 2018 Anal. Chim. Acta, DOI: 10.1016/j.aca.2017.12.034
- 13. 13 Do we need all these libraries? Vinaixa, Schymanski, Navarro, Neumann, Salek, Yanes, 2016, TrAC, DOI: 10.1016/j.trac.2015.09.005 o Yes … most libraries still have many unique entries = HMDB, GNPS, MassBank, ReSpect Compound lists provided by: S. Stein, R. Mistrik, Agilent
- 14. 14 Mind the Gap! Frainay, C. et al. (2018) “Mind the Gap: …” Metabolites: http://www.mdpi.com/2218-1989/8/3/51 o Best library to choose depends highly on your dataset • Example: MSforID (https://msforid.com/) is poor for metabolic networks – but great for forensic toxicology!
- 15. 15 Comparability: QTOF v Orbitrap Oberacher et al. 2019, Metabolites, 9(1), 3; https://doi.org/10.3390/metabo9010003 Orbitrap QTOF
- 16. 16 MassBank EU http://massbank.eu/MassBank https://github.com/MassBank/MassBank-data/ >52,800 spectra >40 contributors
- 17. 17 MassBank EU https://github.com/MassBank/MassBank-data; https://github.com/MassBank/MassBank-web/; Rösch et al DOI 10.1021/acs.est.5b05186 http://massbank.eu/MassBank o MassBank.EU was founded late 2012, hosted at UFZ, Leipzig, Germany o >16,000 MS/MS spectra; 1,200 substances from NORMAN members o Thorough Github-based modernization in progress for traceability: o Tentative/unknown/literature spectra (Level Scheme) as SI for publications Schymanski et al DOI: 10.1021/es5002105
- 18. 18 Confidence Levels for Tentative Structures Schymanski, Jeon, Gulde, Fenner, Ruff, Singer & Hollender (2014) ES&T, 48 (4), 2097-2098. DOI: 10.1021/es5002105 o Annotation is the key to communicating information MS, MS2, RT, Reference Std. Level 1: Confirmed structure by reference standard Level 2: Probable structure a) by library spectrum match b) by diagnostic evidence Identification confidence N N N NHNH CH3 CH3 S CH3 OH MS, MS2, Library MS2 MS, MS2, Exp. data Example Minimum data requirements Level 4: Unequivocal molecular formula Level 5: Exact mass of interest C6H5N3O4 192.0757 MS isotope/adduct MS Level 3: Tentative candidate(s) structure, substituent, class MS, MS2, Exp. data
- 19. 19 Creating High-Quality Mass Spectra Stravs, Schymanski, Singer and Hollender, 2013, Journal of Mass Spectrometry, 48, 89–99. DOI: 10.1002/jms.3131 Automatic MS and MS/MS Recalibration and Clean-up Remove interfering peaks Spectral Annotation with - Experimental Details - Compound Information https://github.com/MassBank/RMassBank/ http://bioconductor.org/packages/RMassBank/
- 20. 20 Communicating Mass Spectra for Mixtures Stravs et al. (2013), J. Mass Spectrom, 48(1):89-99. DOI: 10.1002/jms.3131 OHSO O CH3 O OH m n SPA-9C m+n=6 Formulas: http://sourceforge.net/projects/genform/ Meringer et al, 2011, MATCH 65, 259-290 Data: Schymanski et al. 2014, ES&T, 48: 1811-1818. DOI: 10.1021/es4044374 Chromatography and MS/MS Annotation Literature: LIT00034,35 Sample: ETS00002 Standard: ETS00016,17,19,20 https://github.com/MassBank/RMassBank/
- 21. 21 Suspect Screening: Different Approaches Target List Suspect List HPLC separation and HR-MS/MS TARGET ANALYSIS SUSPECT SCREENING Targets found Suspects found Confirmation and quantification of compounds present Candidate selection (retention time, MS/MS, calculated properties) Sampling extraction (SPE) HPLC separation HR-MS/MS o Search in mass spectral libraries o Screen for predicted transformation products of known parent compounds o Look for “well known” substances without reference standards o Screen for known homologue series
- 22. 22 Suspect Screening: Benzotriazole TPs Huntscha et al. 2014, ES&T, 48(8), 4435-4443. DOI: 10.1021/es405694z 28 Suspects HPLC separation and HR-MS/MS SUSPECT SCREENING 11 masses for 6 suspect formulas 7 with MS/MS 1 reference std. 1 TP confirmed 1 TP “likely”, no std. [UM-PPS] ↓ Eawag-PPS ↓ [enviPath]
- 23. 23 Suspect Screening: Benzotriazole TPs Huntscha et al. 2014, ES&T, 48(8), 4435-4443. DOI: 10.1021/es405694z 28 Suspects HPLC separation and HR-MS/MS SUSPECT SCREENING 11 masses for 6 suspect formulas 7 with MS/MS 1 reference std. 1 TP confirmed 1 TP “likely”, no std. [UM-PPS] ↓ Eawag-PPS ↓ [enviPath] N N N H O OH N N N H O OH - Predicted with Eawag-PPS - No standard - Not in databases (at that time) - Confirmed with reference std. - Observed in WWTP effluents
- 24. 24 Suspect Screening: Benzotriazole TPs Huntscha et al. 2014, ES&T, 48(8), 4435-4443. DOI: 10.1021/es405694z 1H-BT .eu
- 25. 25 Suspect Screening: Different Approaches Target List Suspect List HPLC separation and HR-MS/MS TARGET ANALYSIS SUSPECT SCREENING Targets found Suspects found Confirmation and quantification of compounds present Candidate selection (retention time, MS/MS, calculated properties) Sampling extraction (SPE) HPLC separation HR-MS/MS o Search in mass spectral libraries o Screen for predicted transformation products of known parent compounds o Look for “well known” substances without reference standards o Screen for known homologue series
- 26. 26 (European) Environmental Community (subset!) Schymanski et al. 2015, ABC, DOI: 10.1007/s00216-015-8681-7 Croatian Water RWS Specialist Knowledge Highly Disjointed
- 27. 27 European (World-)Wide Exchange of Suspects Schymanski et al. 2015, ABC, DOI: 10.1007/s00216-015-8681-7 NORMAN Suspect List Exchange: https://www.norman-network.com/?q=suspect-list-exchange
- 28. 28 NORMAN Suspect List Exchange https://www.norman-network.com/?q=suspect-list-exchange Schymanski, Aalizadeh et al. in prep; https://www.researchgate.net/project/Supporting-Mass-Spectrometry-Through-Cheminformatics ReferencesFull Lists
- 29. 29 NORMAN SusDat (merged list) https://www.norman-network.com/nds/susdat/ https://comptox.epa.gov/dashboard/chemical_lists/susdat Schymanski, Aalizadeh et al. in prep; https://www.researchgate.net/project/Supporting-Mass-Spectrometry-Through-Cheminformatics
- 30. 30 o Now >50 lists available online … from small to large! • Specialist collections (e.g. NormaNEWS) to large market lists • Integrated into the CompTox Chemistry Dashboard NORMAN Suspect Exchange Lists
- 31. 31 RECAP: Target Analysis: Status Quo (>364 targets) Schymanski et al. (2014), ES&T, 48: 1811-1818. DOI: 10.1021/es4044374 Target List HPLC separation and HR-MS/MS TARGET ANALYSIS Targets found Confirmation and quantification of compounds present Sampling extraction (SPE) HPLC separation HR-MS/MS
- 32. 32 What about Non-Target Screening? Target List Suspect List (no prior information) HPLC separation and HR-MS/MS TARGET ANALYSIS SUSPECT SCREENING NON-TARGET SCREENING Targets found Suspects found Masses of interest (Molecular formula) DATABASE SEARCH STRUCTURE GENERATION Confirmation and quantification of compounds present Candidate selection (retention time, MS/MS, calculated properties) Sampling extraction (SPE) HPLC separation HR-MS/MS Number of compounds
- 33. 33 Target List Suspect List (e.g. NORMAN, LMC, Eawag-PPS, ReSOLUTION) Componentization (nontarget) TARGET ANALYSIS SUSPECT SCREENING NON-TARGET SCREENING (enviMass, vendor software) Gather evidence (nontarget, ReSOLUTION, RMassBank) Masses of interest Molecular formula determination (enviPat, GenForm) Non-target identification (MetFrag2.3, ReSOLUTION) Sampling extraction (SPE) HPLC separation HR-MS/MS Detection of blank/blind/noise/internal standards; time trend analysis (enviMass) Conversion (Proteowizard) and Peak Picking (enviPick, xcms, MZmine, …) Prioritization (enviMass) MS/MS Extraction (RMassBank) Interpretation, confirmation, peak inventory, confidence and reporting
- 34. 34 MetFrag Relaunched! Ruttkies, Schymanski, Wolf, Hollender, Neumann (2016) J. Cheminf., 2016, DOI: 10.1186/s13321-016-0115-9 Status: 2010 => 2019 5 ppm 0.001 Da mz [M-H]- 213.9637 ChemSpider or PubChem± 5 ppm RT: 4.54 min 355 InChI/RTs References External Refs Data Sources RSC Count PubMed Count Suspect Lists MS/MS 134.0054 339689 150.0001 77271 213.9607 632466 Elements: C,N,S S OO OH
- 35. 35 https://msbi.ipb-halle.de/MetFragBeta/ ; https://comptox.epa.gov/dashboard/ ; https://massbank.eu ; http://normandata.eu/ Combined evidence clearly highlights potential neurotoxicant among chemical candidates Connecting Resources in MetFrag
- 36. 36 MetFrag Relaunched! Ruttkies, Schymanski, Wolf, Hollender, Neumann (2016) J. Cheminf., 2016, DOI: 10.1186/s13321-016-0115-9 Try with the Web Interface: http://msbi.ipb-halle.de/MetFragBeta/
- 37. 37 Ruttkies, Schymanski, Wolf, Hollender, Neumann (2016) J. Cheminf., 2016, DOI: 10.1186/s13321-016-0115-9 Try with the Web Interface: http://msbi.ipb-halle.de/MetFragBeta/ MetFrag Relaunched!
- 38. 38 State of the Art in Small Molecule Identification Schymanski et al, 2017, J Cheminf., DOI: 10.1186/s13321-017-0207-1 www.casmi-contest.org Metadata is critical to improving annotation of known unknowns!
- 39. 39 Connecting and Enhancing Open Resources https://www.slideshare.net/EmmaSchymanski/small-molecules-in-big-data-analytica-munich o Sharing knowledge is a win-win situation 2014 2015: found in waters across Europe 2016: 1 datapoint cross-annotates 3072 in GNPS Hits in GNPS MassIVE datasets: Surfactants: http://goo.gl/7sY9Pf 2017: Early-Warning System is born 2018: Highlighted in Science
- 40. 40 NORMAN Digital Sample Freezing Platform “Live” retrospective screening of known and unknown chemicals in European samples (various matrices) http://norman-data.eu/ and Alygizakis et al, submitted.
- 41. 41 Interactive heatmap available at http://norman-data.eu/NORMAN-REACH NORMAN Digital Sample Freezing Platform Retrospective screening of REACH chemicals in Black Sea samples (various matrices)
- 42. 42 Real-time Monitoring of the Rhine River Hollender, Schymanski, Singer & Ferguson, 2018, ES&T Feature, 51:20, 11505-11512. DOI: 10.1021/acs.est.7b02184 Previously unknown chemicals detected due to “stand-out” patterns
- 43. 43 NORMAN Digital Sample Freezing Platform Future work: use results of unknowns to drive prioritization efforts http://norman-data.eu/ and Alygizakis et al, submitted
- 44. 44 We still have many unknowns … (l) Data from Schymanski et al 2014, ES&T DOI: 10.1021/es4044374. (r) E. coli data provided by N. Zamboni, IMSB, ETH Zürich. Environment Cells
- 45. 45 NORMAN Suspects don’t all have structures!
- 46. 46 Homologous Series Detection M. Loos & H Singer, 2017. J. Cheminf. DOI: 10.1186/s13321-017-0197-z & Schymanski et al. 2014, ES&T DOI: 10.1021/es4044374 http://www.envihomolog.eawag.ch/ Search for discrete mass differences S OO OH CH3 CH3 m n C9H19 O O S O O OHm
- 47. 47 Towards high throughput MS screening of UVCBs o https://github.com/schymane/RChemMass/
- 48. 48 Supporting Evidence for Homologues Stravs et al. (2013), J. Mass Spectrom, 48(1):89-99. DOI: 10.1002/jms.3131 OHSO O CH3 O OH m n SPA-9C m+n=6 Formulas: http://sourceforge.net/projects/genform/ Meringer et al, 2011, MATCH 65, 259-290 Data: Schymanski et al. 2014, ES&T, 48: 1811-1818. DOI: 10.1021/es4044374 Chromatography and MS/MS Annotation Literature: LIT00034,35 Sample: ETS00002 Standard: ETS00016,17,19,20 https://github.com/MassBank/RMassBank/
- 49. 49 Cross-Linking Homologues in the Dashboard Schymanski, Grulke, Williams et al, in prep. & Williams et al. 2017 J. Cheminformatics 9:61 DOI: 10.1186/s13321-017-0247-6 https://comptox.epa.gov/dashboard/chemical_lists/eawagsurf
- 50. 50 Homologous Series in Biological Matrices Lipid extract of Mycobacterium smegmatis C23F48O7 +CF2 Schymanski & Zamboni … random data exploration …
- 51. 51 Exchanging Knowledge … Open Science Helps! We need to be able to find and annotate the unexpected! C23F48O7 +CF2 Schymanski & Zamboni … random data exploration …
- 52. 52 Exchanging Knowledge … Open Science Helps! We need to be able to find and annotate the unexpected!
- 53. 53 Exchanging data reveals things we never expected! Schymanski & Zamboni … random data exploration … o Lipid extract of Mycobacterium smegmatis C23F48O7 +CF2 DTXSID70880513DTXSID70880513
- 54. 54 Community Challenges … and Solutions Data: Schymanski et al 2014, DOI: 10.1021/es4044374; https://www.slideshare.net/EmmaSchymanski/small-molecules-in-big-data-analytica-munich High resolution mass spectrometry AND connecting chemical knowledge
- 55. 55 McEachran et al. 2018, DOI: 10.1186/s13321-018-0299-2; Schymanski & Williams, 2017 ES&T DOI: 10.1021/acs.est.7b01908 “MS-ready” Form for MetaData in MetFrag
- 56. 56 “MS-ready” Form for MetaData in MetFrag
- 57. 57 Future MetaData: Topic-Specific Reference Counts Schymanski, Baker, Williams, Singh et al. submitted. Excel macro: https://figshare.com/s/824f6606644f474c7288 https://comptox.epa.gov/dashboard/chemical_lists/litminedneuro
- 58. 58 Take Home Messages Unknowns and High Resolution Mass Spectrometry o Over 60 % of HR-MS peaks are potentially relevant but unknown Environment Cells
- 59. 59 Take Home Messages o Over 60 % of HR-MS peaks are potentially relevant but unknown o Annotating unknowns requires data and evidence from many different sources o Many excellent workflows available to collate this information o Incorporation of all available metadata is critical to success! o E.g. MetFrag has greatly improved the speed and success of tentative identification of “known unknowns”: 15 % => 89 % Ranked Number 1 o https://ipb-halle.github.io/MetFrag/ Unknowns and High Resolution Mass Spectrometry
- 60. 60 Take Home Messages o Over 60 % of HR-MS peaks are potentially relevant but unknown o Annotating unknowns requires data and evidence from many different sources o Exchange expert knowledge worldwide o Community efforts contribute greatly to improved cross-annotation o Information in the public domain helps everyone! o You never know when it will help you Unknowns and High Resolution Mass Spectrometry Schymanski et al. 2015, ABC, DOI: 10.1007/s00216-015-8681-7; Alygizakis et al. 2018 ES&T, DOI: 10.1021/acs.est.8b00365
- 61. 61 Acknowledgements emma.schymanski@uni.lu Further Information: https://massbank.eu/MassBank/ https://ipb-halle.github.io/MetFrag/ https://comptox.epa.gov/dashboard/ http://www.norman-network.com/?q=node/236 https://wwwen.uni.lu/lcsb/research/ environmental_cheminformatics .eu EU Grant 603437
- 62. 62






Be the first to comment