Join GitHub today
GitHub is home to over 36 million developers working together to host and review code, manage projects, and build software together.
Sign upAdd DTXSIDs to all MassBank records with InChIKey match #66
Comments
This was referenced May 7, 2019
This comment has been minimized.
This comment has been minimized.
|
I will take care of this. And I would like to give a short update about a related topic: I curated all records with any structural information available to contain proper InChI and InChI-Keys. There are just 900 records left which dont have structural information, just chemical names. |
This comment has been minimized.
This comment has been minimized.
|
Great! |
This comment has been minimized.
This comment has been minimized.
|
noStructure.txt |
This comment has been minimized.
This comment has been minimized.
|
Oh interesting ... so the EawagAdditional are ones that almost certainly don't have a structure because they are tenative records ... but I see a lot from BS, Fac_Eng_Univ_Tokyo (major culprit) and even IPB Halle! @sneumann should be able to comment about the latter ... do you see a systematic issue (one critical identifier missing that we could fill in with other information available) with BS and Fac_Eng_Univ_Tokyo? |
This comment has been minimized.
This comment has been minimized.
|
There are roughly 60 records with other database identifier, like CAS, which I could use to retrieve proper chemical information. The remaining records have only chemical names. Needs manual lookup and might be unsuccessful in some cases. This will take some time... Different topic: |
This comment has been minimized.
This comment has been minimized.
|
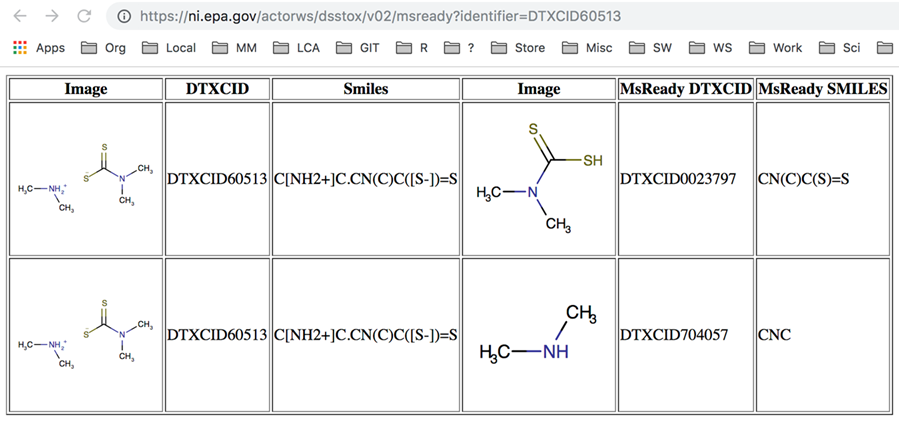
C = compound/chemical and S = substance. The "C" entries are the unique chemical (~~ "MS-ready" forms (put simply)) and the "S" entries are the official database entry. Check out infoboxes here (@ChemConnector note inconsistencies in the DTXCID!) |
This comment has been minimized.
This comment has been minimized.
|
Sorry, didnt understand this concept. On pubchem we have SID which is something like the label on a bottle with chemicals and could potentially be a mixture and we have CID which is a uniqe compound which is represented by exactly one formula(like you would draw on a paper). Thats why more questions: |
This comment has been minimized.
This comment has been minimized.
|
As far as I'm aware it's a one DTXSID per InChIKey. The service should return us one DTXSID for one InChIKey request and this is what @ChemConnector asked us to do, use InChIKey to DTXSID to add these identifiers to MassBank .. (therefore I'm assuming this is the most robust way in his opinion and from my experience, I'd agree) One DTXSID may have multiple DTXCIDs associated with it. It's a bit different to the PubChem construct. imho we should not yet try mapping on DTXCIDs as they don't have the full functionality associated with them like the DTXSIDs, until recently they were hidden entirely. Some examples: https://comptox.epa.gov/dashboard/dsstoxdb/results?search=DTXSID10858175 |
This comment has been minimized.
This comment has been minimized.
|
I have created a program which can add these identifier with the help of the InChI-key to DTXSID resolver and have processed all records. We have now 39962 outlinks in place. This program can be executed on all new records and also on a regular basis on the existing records. I think this one can be closed. |
 meier-rene
closed this
meier-rene
closed this
May 14, 2019
This comment has been minimized.
This comment has been minimized.
|
Reopen until #68 is solved. |
 meier-rene
reopened this
meier-rene
reopened this
May 16, 2019
This comment has been minimized.
This comment has been minimized.
This was referenced Jun 18, 2019
This comment has been minimized.
This comment has been minimized.
|
Note that if the cause of the problem is the web services return also up to Level 6, if the "curation level" would be in the data retrieved, we could proactively fix our end by only including DTXSIDs if the level is 5 or lower. I can't see that this information is included yet tho, just following the links above - although I thought this was part of the plan @ChemConnector ? |



schymane commentedMay 7, 2019
@meier-rene @Treutler the EPA have set up a basic service that should allow retrieval of DTXSIDs by InChIKey, can you look into implementing this on the database end to add DTXSIDs to all records with matching entries for now, I will post a separate issue to get this into RMassBank and linked up in MassBank-web.
It's already in our Record format as
CH$LINK: COMPTOX DTXSID50274017
(https://github.com/MassBank/MassBank-web/blob/master/Documentation/MassBankRecordFormat.md)
https://actorws.epa.gov/actorws/chemIdentifier/v01/resolve?identifier=IKHGUXGNUITLKF-UHFFFAOYSA-N
https://actorws.epa.gov/actorws/chemIdentifier/v01/resolve.json?identifier=IKHGUXGNUITLKF-UHFFFAOYSA-N
https://actorws.epa.gov/actorws/chemIdentifier/v01/resolve.xml?identifier=IKHGUXGNUITLKF-UHFFFAOYSA-N
Any feedback re service to @ChemConnector
Thanks!