Join GitHub today
GitHub is home to over 40 million developers working together to host and review code, manage projects, and build software together.
Sign up[REVIEW]: py-pde: A Python package for solving partial differential equations #2158
Comments
This comment has been minimized.
This comment has been minimized.
|
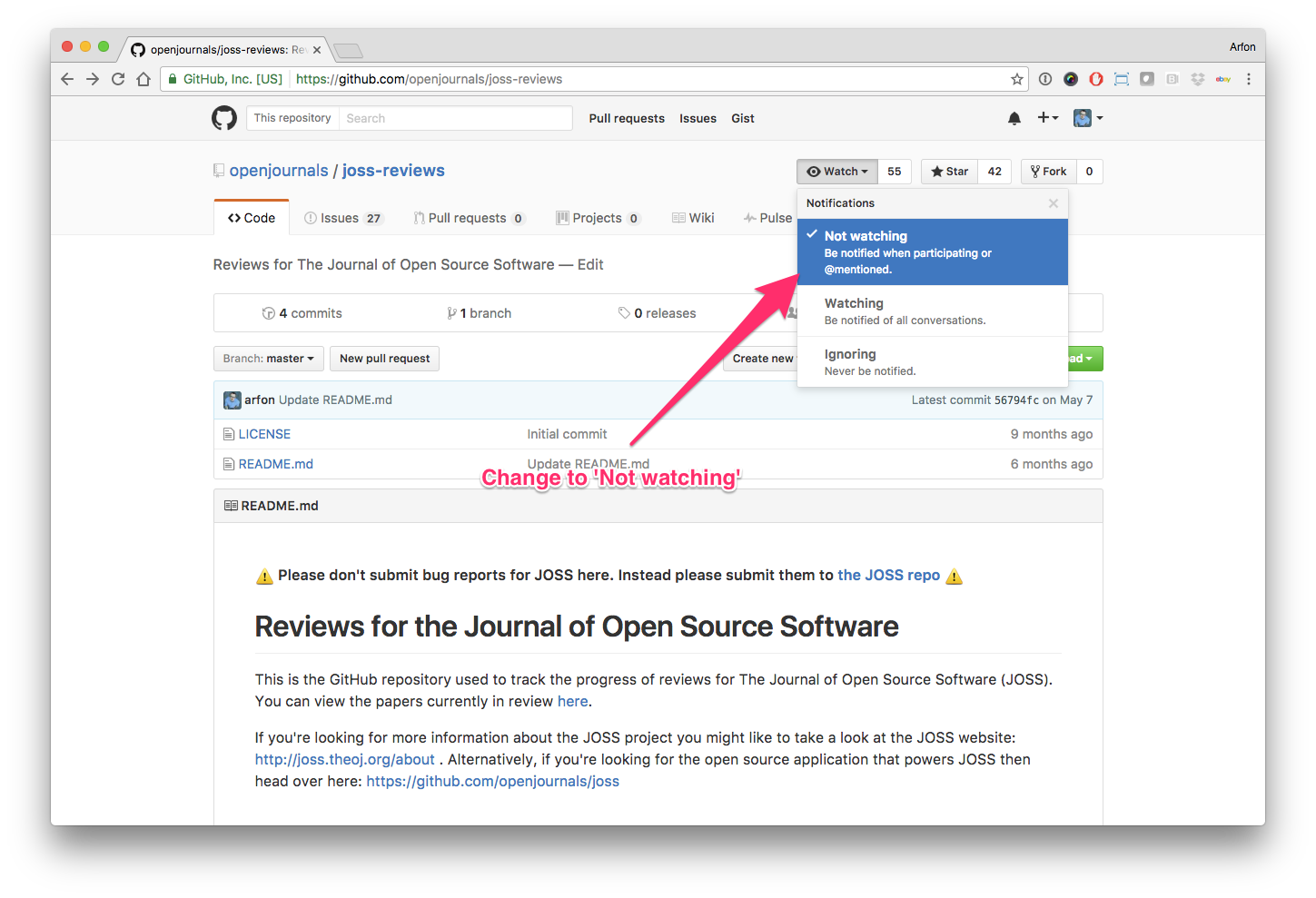
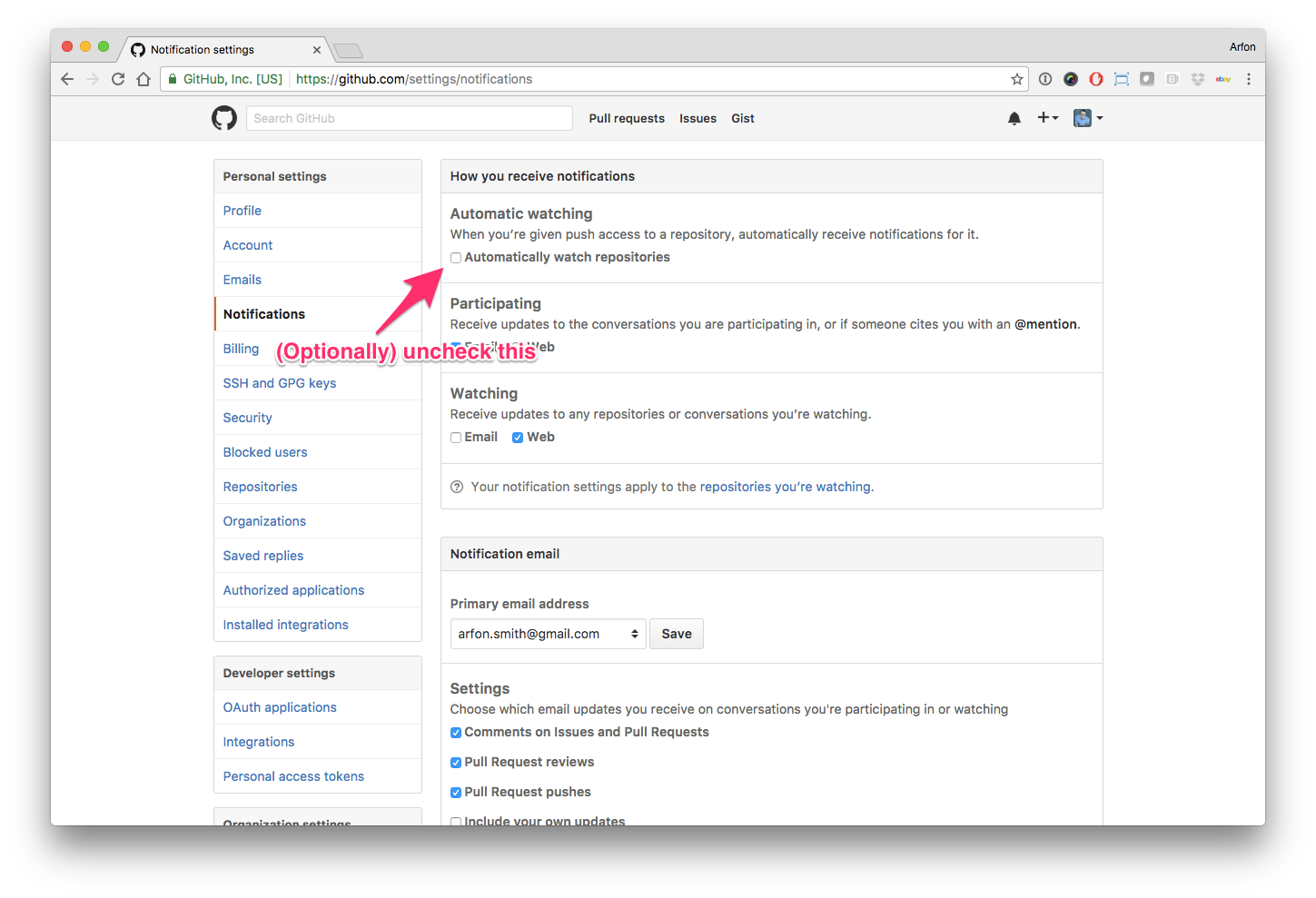
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @celliern, @@mstimberg it looks like you're currently assigned to review this paper If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews To fix this do the following two things:
For a list of things I can do to help you, just type: For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type: |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
PDF failed to compile for issue #2158 with the following error: pandoc-citeproc: reference mstimberg not found Looks like we failed to compile the PDF |
This comment has been minimized.
This comment has been minimized.
|
I think there was an issue with |
This comment has been minimized.
This comment has been minimized.
|
This looks like a problem with a reference in the .md and .bib files - the .md file seems to be trying to reference something that is not in the bib file (or the bib file has a problem that leads to it not being parsed correctly) |
This comment has been minimized.
This comment has been minimized.
|
|
This comment has been minimized.
This comment has been minimized.
|
@whedon generate pdf |
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
@xuanxu I cannot edit the checklist and I did not receive an invite – is this because this is my first review for JOSS and I have to do something else first? Or is it because of the earlier |
This comment has been minimized.
This comment has been minimized.
|
@mstimberg I guess it's the previous |
This comment has been minimized.
This comment has been minimized.
|
@whedon re-invite @mstimberg as reviewer |
This comment has been minimized.
This comment has been minimized.
|
OK, the reviewer has been re-invited. @mstimberg please accept the invite by clicking this link: https://github.com/openjournals/joss-reviews/invitations |
This comment has been minimized.
This comment has been minimized.
|
Thanks @xuanxu, it's working now |
This comment has been minimized.
This comment has been minimized.
|
First, I wanted to say that I'm impressed ! The library is clean and well organized and I will definitively add it in my research toolbox. It also fit well the Python scientific ecosystem, especially compared with my work () that has not be designed to easily write tensor-based PDE equation. This is a welcome approach as this is a very common task among a wide range of scientific topics. The features offered by py-pde are multiple and the dependencies despite very narrowed mandatory dependencies (all of them being in the standard scientific stack and shipped with distributions like Anaconda). This should guarantee an easy deployment on clusters. All tests have run with a fresh conda environment with py=3.6 on my computer. I still have to play a bit with it to ensure the functional claims of the software. During this time, I have some optional (but welcome) improvement that I could suggest : Packaging improvementThe optional dependencies could be listed in the Documentation improvementSome information are available in the paper may be added also in the documentation: the method used for the operator discretization (finite difference), the temporal solvers available for the explicit as well as for the implicit ones. In general, a "advanced user documentation" could highly improve the adoption of the software : more detail on how to implement custom PDE (with or without numba), some info on the internals (discretization, time-steps choice), and if / how the library can be extended to fit more specific needs. Similarly, there is no in-between the very short overview and trivial example and the full API documentation (which is welcome but hard to decipher). Some doc-strings lack of details : the allowed boundary conditions are well described in the raised ValueError, but not in the doc-string itself. There is more examples available on the repository than shown in the documentation : using Sphinx-Gallery, these examples could easily be integrated in the documentation. The software seems to embed a benchmark tool : it could be nice to have a summary of the performance of implemented models with and without numba and with and without parallelization. Community guidelinesIt could be nice to add a Code of Conduct (see the scipy one for an extended example, and here for a standard one that you can easily adapt for your repository). |
This comment has been minimized.
This comment has been minimized.
|
Dear authors and reviewers We wanted to notify you that in light of the current COVID-19 pandemic, JOSS has decided to suspend submission of new manuscripts and to handle existing manuscripts (such as this one) on a "best efforts basis". We understand that you may need to attend to more pressing issues than completing a review or updating a repository in response to a review. If this is the case, a quick note indicating that you need to put a "pause" on your involvement with a review would be appreciated but is not required. Thanks in advance for your understanding. Arfon Smith, Editor in Chief, on behalf of the JOSS editorial team. |
This comment has been minimized.
This comment has been minimized.
|
Dear @celliern, thank you very much for your suggestions! I managed to include most of your points in the revised code. Thank you especially for suggesting the example gallery – I didn't know about this, but I think it helps showcasing the features of the package tremendously. I'm looking forward to the remaining comments, but I obviously understand if this will now take much longer than usual. Stay safe and healthy! |
This comment has been minimized.
This comment has been minimized.
|
@david-zwicker Nice your gallery is wonderful ! Two minor points :
As a side suggestion (not blocking for the publication) : you will have a strong limitation on hyperbolic equations. This can be (partially) mitigated by introducing "advection" operator that could either implement really simple Gudunov finite volume scheme or upwind finite difference scheme. This will extend the scope of the software. Stay safe and healthy as well ! |
This comment has been minimized.
This comment has been minimized.
|
For the paper:
Otherwise, the paper look fine. I will recommand the paper / software for publication once these different point are fixed. You have written a nice scientic tool. Nice work ! |
This comment has been minimized.
This comment has been minimized.
|
I join @celliern in congratulating you to your software, this is really an excellent tool and I have rarely seen that much attention to code quality in scientific software (type annotations, etc.)! I have reported two minor issues (zwicker-group/py-pde#2, zwicker-group/py-pde#3) and three suggestions (zwicker-group/py-pde#4, zwicker-group/py-pde#5, zwicker-group/py-pde#5) directly in the Fixing the typos in the article is the only real blocker for me – if I wanted to be really nitpicky, I could also mention that the capitalization in the references list is somewhat inconsistent (e.g. "Python" vs. "python"). Pending these minor issues I can easily recommend this paper for publication. |
This comment has been minimized.
This comment has been minimized.
Please do be nitpicky - we need to fix all of these before publication :) |
This comment has been minimized.
This comment has been minimized.
|
OK @david-zwicker, everything looks good, here are the next steps:
Once you do that please report here the version number and archive DOI |
This comment has been minimized.
This comment has been minimized.
|
I just published the latest package on github and Zenodo:
The GitHub version is v0.5 and the DOI is https://doi.org/10.5281/zenodo.3739300
… On Apr 3, 2020, at 14:32, Juanjo Bazán ***@***.***> wrote:
OK @david-zwicker, everything looks good, here are the next steps:
• Please release a new tagged version from the current master so it includes all the changes made during the review process
• Then archive that latest release in Zenodo
• Check the Zenodo deposit has the correct metadata: title and author name should match the paper; you may also add your ORCID.
Once you do that please report here the version number and archive DOI
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub, or unsubscribe.
|
This comment has been minimized.
This comment has been minimized.
|
@whedon set v0.5 as version |
This comment has been minimized.
This comment has been minimized.
|
OK. v0.5 is the version. |
This comment has been minimized.
This comment has been minimized.
|
@whedon set 10.5281/zenodo.3739300 as archive |
This comment has been minimized.
This comment has been minimized.
|
OK. 10.5281/zenodo.3739300 is the archive. |
This comment has been minimized.
This comment has been minimized.
|
Thanks @mstimberg and @celliern for your reviews! |
This comment has been minimized.
This comment has been minimized.
|
@whedon accept |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
Check final proof If the paper PDF and Crossref deposit XML look good in openjournals/joss-papers#1405, then you can now move forward with accepting the submission by compiling again with the flag |
This comment has been minimized.
This comment has been minimized.
|
Thanks @xuanxu - I'll take over |
This comment has been minimized.
This comment has been minimized.
|
|
This comment has been minimized.
This comment has been minimized.
|
@whedon accept |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
Check final proof If the paper PDF and Crossref deposit XML look good in openjournals/joss-papers#1406, then you can now move forward with accepting the submission by compiling again with the flag |
This comment has been minimized.
This comment has been minimized.
|
@whedon accept deposit=true |
This comment has been minimized.
This comment has been minimized.
|
This comment has been minimized.
This comment has been minimized.
|
|
This comment has been minimized.
This comment has been minimized.
|
Here's what you must now do:
Any issues? notify your editorial technical team... |
This comment has been minimized.
This comment has been minimized.
|
Thanks to @celliern & @mstimberg for reviewing, and @xuanxu for editing! Congratulations to @david-zwicker! |
This comment has been minimized.
This comment has been minimized.
|
If you would like to include a link to your paper from your README use the following code snippets: This is how it will look in your documentation: We need your help! Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
|
This comment has been minimized.
This comment has been minimized.
|
Thank you everyone for the very smooth process and for your numerous suggestions that surely improved the quality of the code, the documentation, and the paper. There is some small issue still: I cannot see the final pdf of the paper and I actually receive a 404 when I want to download it. Is this something that would just take a while until it is finally there? |
This comment has been minimized.
This comment has been minimized.
|
There's likely a caching issue on your side - it works fine for me |
Submitting author: @david-zwicker (David Zwicker)
Repository: https://github.com/zwicker-group/py-pde
Version: v0.5
Editor: @xuanxu
Reviewer: @celliern, @mstimberg
Archive: 10.5281/zenodo.3739300
Status
Status badge code:
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@celliern & @@mstimberg, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @xuanxu know.
Review checklist for @celliern
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
Review checklist for @mstimberg
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper